Jay Yang
Singer Instruments, Roadwater, Somerset, UK, TA23 0RE
Summary
Tetrad dissection is a vital tool for yeast geneticists. It is a method by which sporulating yeast cells are teased apart, and the individual cells isolated. Specially designed micromanipulation tools are used to carefully separate the cells of interest, which are then transferred to a new growth medium for further study.
Historically, tetrad dissection was a cumbersome and difficult procedure involving a high level of manual dexterity and skill, acquired through years of practice. In recent times the technique has become much easier, largely due to the development of easy-to-use micromanipulation systems.
Principles
Two yeast strains of interest – one strain MATa and one MATα – are mated producing diploid cells (containing two sets of chromosomes).
These cells are transferred to a low-nutrient medium, which stimulates each diploid cell to undergo the process of sporulation, resulting in a tetrad of four haploid spores (containing half the usual complement of chromosomes) encased in a small membrane.
(Figure1)
The tetrads are subjected to a carefully timed enzymatic digestion, sufficient to break down the protective membrane surrounding the spores but not long enough to disrupt and cause the whole tetrad to fall apart.
The digested tetrads are transferred onto an agar surface on a microscopic platform. Tetrads are selected, picked, and the individual spores are pulled apart using a micromanipulator device that controls the movement of a fine glass needle. (Figure 2)
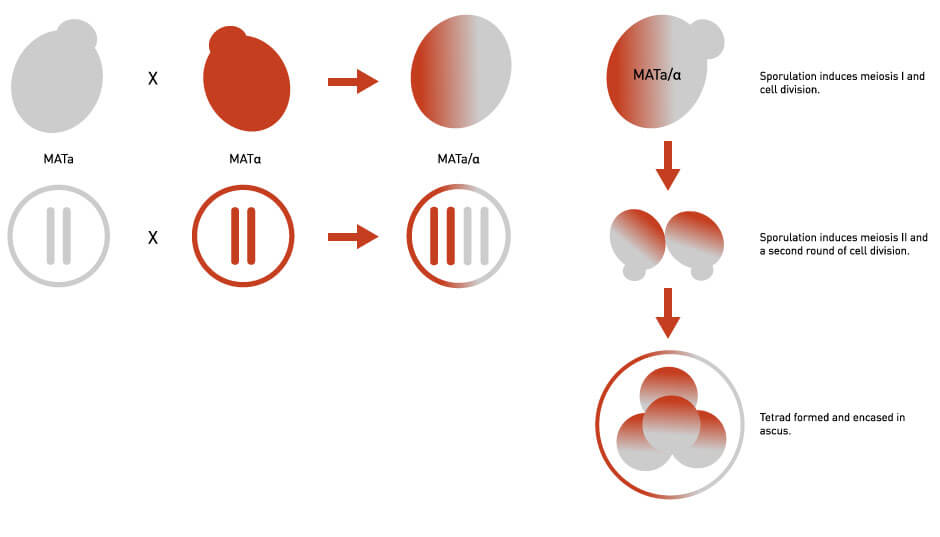
Figure 1: Diagrams Illustrating Yeast Mating and Sporulation. See text for details.
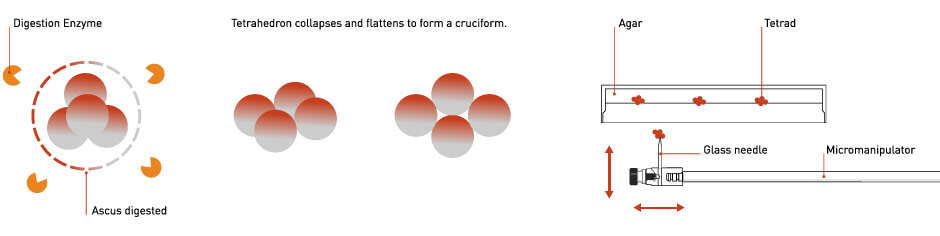
Figure 2: Diagrams Illustrating Tetrad Digestion and Dissection. See text for details
Theory
When yeast undergoes normal cell division the progeny are genetically identical to the parental cells. However, it is sometimes desirable to cross different strains of yeast in order to study genetic linkages (how one gene can produce different phenotypes), produce new strains with unique genetic combinations, or engineer mutations into a strain.
In order to achieve this, yeast strains must be mated. The two mating types, MATa and MATα, which possess the genetic qualities to be studied fuse to form cells containing both sets of chromosomes, and are then induced to sporulate by removing nutrients from their growth media. During this sporulation cells undergo two rounds of cell division, resulting in four spores in which the chromosomes have been distributed.
These four spores must be physically separated in order to determine their new genetic make-up and isolate desirable cells for further study and expansion.
Digestion
The protective membrane, the ascus, which contains the four spores of the tetrad is then digested using an enzyme that breaks down the ascus, for example, zymolase. Best practice is to perform parallel digestions at several time-points in order to obtain the best tetrads.
The tetrads must be sufficiently digested so the ascus is dissolved, but the four spores do not fall apart prior to dissection. A properly digested tetrad will collapse from a tetrahedron into a flattened cruciform shape. A protocol for tetrad digestion can be found here.
Dissection
Tetrads are plated onto an agar dish or agar-coated glass slide. This is inverted onto a specialized microscope workstation that is designed for yeast tetrad dissection. These microscopes incorporate a micromanipulator device so the user can pick tetrads from the agar surface, replate the tetrads in newly defined positions on the plate and separate the four spores as desired. An example of a semi-automated dissection microscope is the Singer MSM 400.
Application in Genetic Linkage
Tetrad dissection has been extensively used in genetic linkage analysis to determine how close two mutations are on a chromosome (1). If the parental cells contain mutations in two different genes, the tetrad can segregate these genes as parental ditype (PD), non-parental ditype (NPD), and tetrad type (T) (Figure 3). Parental ditype (PD) is a tetrad that exhibits two genotypes, both of which are parental. Non-parental ditype (NPD) is a tetrad that exhibits two recombinant genotypes that are different from both parents. Tetrad type (T) is a tetrad that exhibits four different genotypes, two parental and two recombinant.
The frequency of PD, NPD, and T can be used to determine the distance, or linkage, between two genes. When two genes are linked, or physically close together on a chromosome, the frequency of PD will be much greater than NPD. On the other hand, when two genes are on different chromosomes or when two genes are widely separated on the same chromosome, there will be no linkage between the genes and the ratio of PD: NPD: T will be 1: 1: 4.
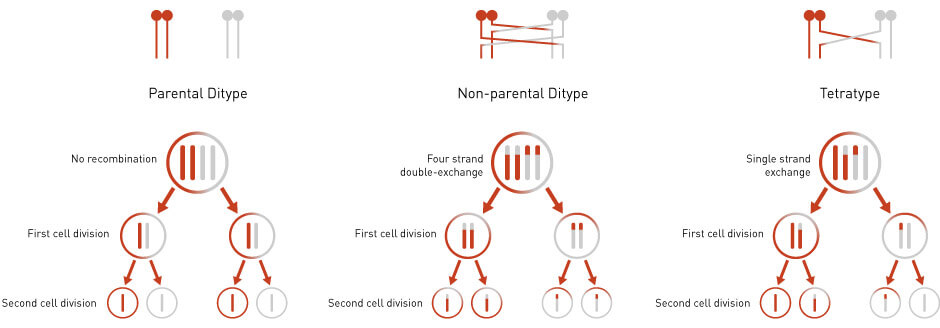
Figure 3: An Example of Tetrad Analysis. See text for details
Other Applications for Tetrad Dissection
In addition to linkage analysis, tetrad dissection is necessary to determine if a mutation corresponds to an alternation at a single locus, to construct new mutant strains by combining two parental gene mutations, and to investigate genetic interactions. For more information on how to perform tetrad dissection, please click here to view the MSM 400.
Yeast Tetrad Dissection Applications
Finding the sources of missing heritability in a yeast cross Abstract Bloom JS, Ehrenreich IM, Loo WT, Lite T-LV, Kruglyak L
For many traits, including susceptibility to common diseases in humans, causal loci uncovered by genetic-mapping studies explain only a minority of the heritable contribution to trait variation. Multiple explanations for this ‘missing heritability’ have been proposed. Here we use a large cross between two yeast strains to accurately estimate different sources of heritable variation for 46 quantitative traits, and to detect underlying loci with high statistical power. We find that the detected loci explain nearly the entire additive contribution to heritable variation for the traits studied. We also show that the contribution to heritability of gene-gene interactions varies among traits, from near zero to approximately 50 per cent. Detected two-locus interactions explain only a minority of this contribution. These results substantially advance our understanding of the missing heritability problem and have important implications for future studies of complex and quantitative traits.
Reference
Sherman F. Getting started with yeast. Meth Enzymol. 2002;350:3–41.

