Jose E. Aguiar-Cervera1,2 and Oliver Severn2
1 The University of Manchester, Oxford Rd, Manchester M13 9PL
2 Singer Instruments, Roadwater, Watchet TA23 0RE
Author: Prof. Daniela Delneri, FRSB, Chair in Evolutionary Genomics, Manchester Institute of Biotechnology
Funding: H2020 Marie Skłodowska‐CurieActions,Grant/Award Number: H2020‐MSCA‐ITN‐2017(764364)’
Aim
To determine whether PhenoBooth+ can be used to assess the growth performance of conventional and non-conventional yeast strains.
Introduction
High-throughput techniques have become crucial for the discovery of new strains with potential in the beverage industry. These approaches allow the assessment of critical traits for fermented beverages such as; stress tolerance, viability, vitality, and aromatic compound production in a fast and effective manner. Among all traits, viability and vitality of a certain yeast population greatly determine the performance and the metabolic product production in a fermentation1.
A particular strain might have the genetic information to produce desirable metabolic products, but if the viability of the population does not reach certain levels, it will not be suitable for the brewing industry2. In this study, a new method for measuring the vitality of vast Saccharomyces and non-Saccharomyces yeast libraries has been developed. This will allow the screening thousand-fold libraries of industrially relevant and/or novel yeast isolates in a fast, cost-effective manner.
Materials and methods
The vitality of a library composed by Saccharomyces and non-Saccharomyces yeast strains was evaluated by measuring the colony size over time with PhenoBooth+ (Singer Instruments) on different solidified industrially relevant media at 20 and 25 oC. A first screening was carried out with 24 strains known for being good aroma producers 3, 4, which were randomly distributed onto YPD agar plates in a 384 array format using PIXL (Singer Instruments). After 24 h of growth at 30 oC, the ROTOR HDA (Singer Instruments) was used to replicate the arrays onto solidified industrially relevant media, which included: Synthetic Apple Juice (SAJ), Malt Extract (ME), Apple Juice (AJ) and Wort (WO). The plates were incubated at 20 and 25 oC, and PhenoBooth+ (Singer Instruments) was used to capture images every ~ 10 h, during 210 h (Figure 1.A). A total of 340 images were obtained, and PhenoSuite was used to filter the background and normalise the colony sizes taking in account the plate, column and row means. The normalised dataset was analysed and plotted using the gg2plot R package and GraphpadPrism software.
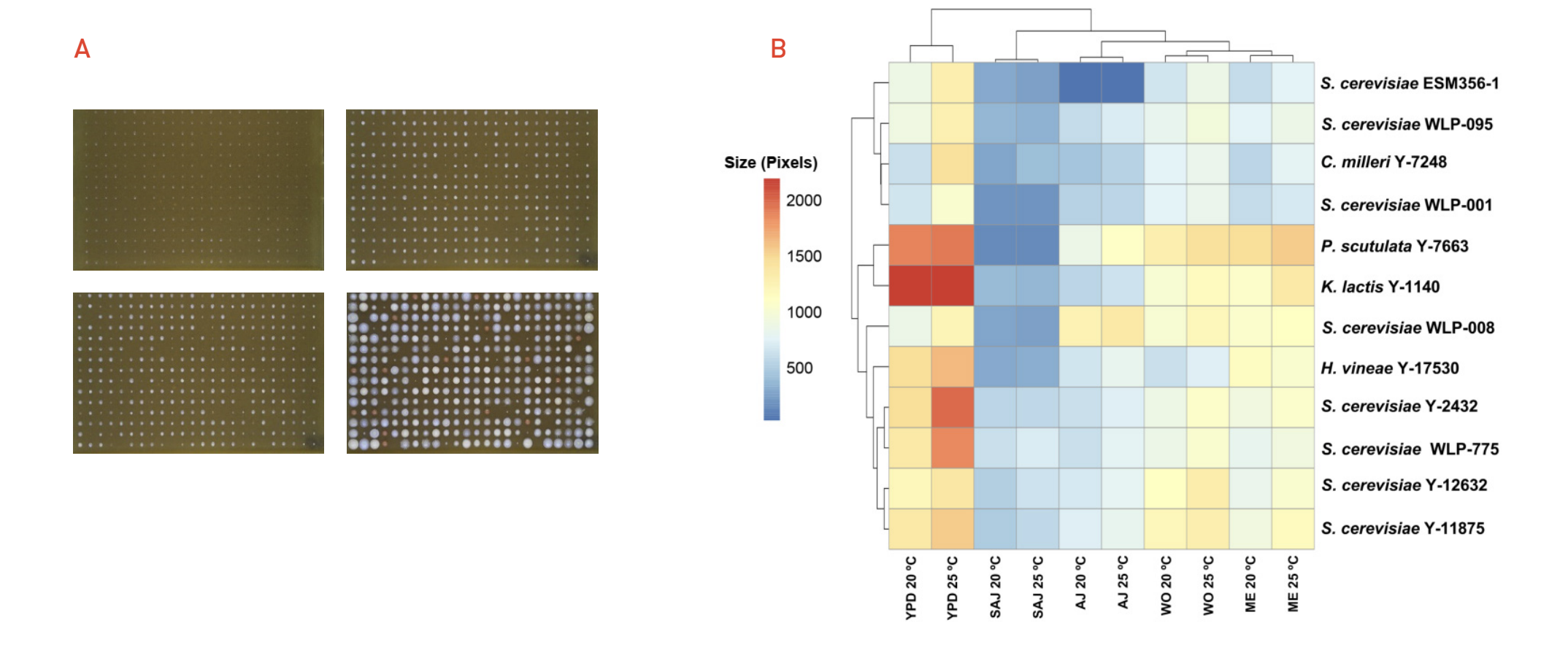
Figure 1. Screening of conventional and non-conventional yeast strains in industrially relevant media. (A) Strains randomly distributed and grown in industrial conditions; Synthetic Apple Juice (SAJ), Malt Extract (ME), Apple Juice (AJ) and Wort (WO); for 210 hours. (B) Average final colony sizes measured in pixels for each temperature and media condition.
Results and Conclusions
Growth overtime (data not shown) and final size measured in pixels (Figure 1.B) provided useful information regarding the capabilities of each strain to grow in a certain condition. For example, the lab strain Saccharomyces cerevisiae ESM356-1 did not show growth in AJ. This is likely to be caused by the fact that this strain is an auxotrophic mutant for various essential amino acids that might not be present in the AJ, and hence the strain is not able to grow on it. Interestingly, two commercially available ale strains: S. cerevisiae WLP-001 and S. cerevisiae WLP-008; showed their bigger colony sizes in AJ medium when a better adaptation to growth in wort or rich media conditions was expected. These strains can be very good candidates for further analysis of fermentation performance and aromatic compound production on liquid apple juice.

