Picking single colonies of Chlamydomonas Reinhardtii from 1536 and 6144-density arrays using The Stinger single colony picker
Kresti Pecani and Frederick R. Cross
Laboratory of Cell Cycle Genetics, The Rockefeller University, New York, NY 10065

We routinely carry out high-throughput screens for temperature-sensitive colonies of Chlamydomonas reinhardtii which would be greatly aided by the use of an automated, single-colony picker capable of accurately picking individual colonies from high-density arrays. We evaluate here the use of The Stinger single colony picker for agar-to-agar transfer of individual colonies of Chlamydomonas from 1536 and 6144 array densities. Specifically, we assess the accuracy with which The Stinger selects the desired colony within the source array, the compatibility of the pin material and transfer speed with the viability of Chlamydomonas cells, and the extent of cross-contamination between cells in the desired colony and those of the surrounding colonies.
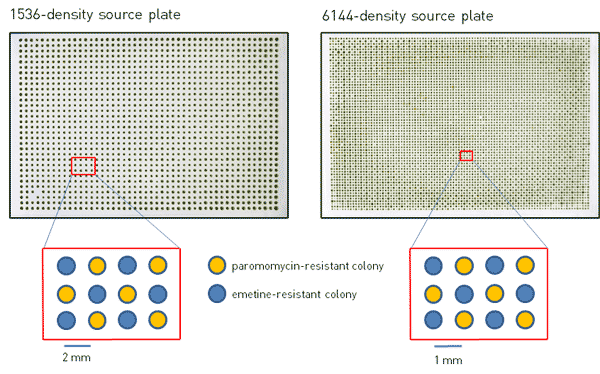
Figure 1. Photographs of 1536- and 6144-density source plates with emetine- and paromomycin-resistant Chlamydomonas colonies arranged in a checkerboard pattern.

We built 1536 and 6144 array source plates using two mutants of Chlamydomonas with drug resistance to either emetine or paromomycin and arranged them in a checkerboard pattern such that each emetine-resistant colony is surrounded by paromomycin-resistant colonies, and vice-versa (Figure 1). The 1536-density source plate was made by using four 384 short-pin RePads to condense two plates containing emetine-resistant cells and two plates containing paromomycin-resistant cells into one 1536 array. The 6144-density source plate was made by the same method using four 1536 RePads onto a flat agar plate lacking a meniscus along the edges. The plates were then incubated at 30˚C for 3 days under constant light (Figure 1). Given the larger footprint of a 6144 array compared to smaller densities, the presence of a meniscus along the edge of the plate would prevent the 6144 RePad from making even contact with all of the agar surfaces, thereby preventing complete transfer of all colonies onto the plate and leaving blank areas within the plate, and/or dense smears on the edges. Therefore, the use of a nearly completely flat plate lacking a meniscus is essential for building and maintaining a 6144-density plate by this method. Our plates are flat to within a <1mm z-tolerance across their surface, which results in a highly accurate transfer using ROTOR+.
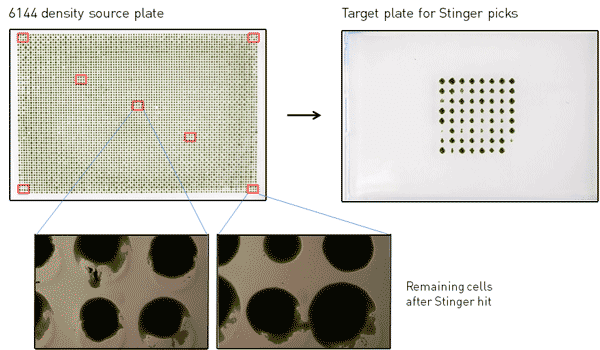
Figure 2. Photograph of 6144-density source plate with approximate regions of colonies picked by The Stinger outlined in red. The plate on the right shows colonies picked by The Stinger after 3 days at 30˚C. Photographs on the bottom show the remaining cells from picked colonies.
The Stinger was used to pick individual colonies from nine 3×3 colony regions from each source plate to a target plate (both at 384 density) (Figure 2). The colonies picked included the four corner colonies, a few colonies along the edges, and various colonies within the array as shown in Figure 2. For both the 1536- and 6144-density source plates, 2x repeat pinning was used with no dry mix and no offset; all other parameters were set to default. Default settings were used for all target plate parameters. Once The Stinger picks were completed, the target plates containing the picked colonies were incubated at 30˚C for 3 days under constant light. When these colonies were grown, we used 384-density RePads to make replicas onto plates containing either emetine or paromomycin (Figure 3). To test for pre-existing cross-contamination in the source plates due to possible irregularities in the transfer from RePads, we also made replicas of the entire source plates immediately following The Stinger picking using 1536/6144-density RePads onto plates containing emetine or paromomycin. In these transfers (data not shown), all colonies transferred with the 1536 or 6144 RePads after The Stinger picks were completely devoid of contamination. This indicates high reproducibility of RePad transfer and shows that plate and pad positions are accurately (error « 1 mm) maintained in separate pinnings days apart.
Following picking with The Stinger, the target plates had viable cells at nearly every pinned position, so the pin material and transfer time is compatible with the viability of Chlamydomonas. There was no detectable cross-contamination of colonies picked from 1536-density: all picked colonies were composed of either solely emetine- or paromomycin-resistant cells, appropriately from their intended spot of origin. However, among all colonies picked from 6144-density, 18% were cross-contaminated with emetine- and paromomycin-resistant cells (average over two 6144-density source plates). If only the first hit in each 3×3 region is considered, the cross-contamination rate was 21%. If the four corner regions are excluded, the cross-contamination rate was 15%, so the likelihood of contamination does not seem to depend on the location of the picked colony. We did observe occasional splashing of cells onto adjacent colonies when The Stinger pin strikes a colony (Figure 2), but the contribution of splashed cells to the total cross-contamination observed must be minimal, since the contamination rate among the first colonies hit in a new region was no better than the overall rate.

Figure 3. indicates the approximate dimension of The Stinger pin surface; it is clear that tremendous accuracy is required at all stages (Stinger pin, plate orientation, lack of agar shrinkage) to avoid the pin striking an adjacent colony. The finding that the 6144 RePad transfer was accurate suggests that some minor misalignment of The Stinger pin is the more likely explanation, but this is not definitive.
The Stinger seems to be accurate and quite effective at transferring Chlamydomonas cells from a 1536-density array, with the high maintenance of viability and no evident contamination from nearby colonies. Picking single colonies from a 6144-density array seems to be more challenging, with a small rate of cross-contamination, and the requirement for a flat, meniscus-free agar plate. Whether this rate of contamination is prohibitive depends on the application.
In one preliminary test, we tried narrow Stinger pins (0.5 rather than 0.75 mm surface), and found 0/12 cross-contaminated colonies at 6144. Therefore, this may provide a solution; this requires further evaluation.

