Case Study
Jay Yang
Singer Instruments, Roadwater, Somerset, UK, TA23 0RE
23/03/2015
Introduction
Mutations are defined as any permanent changes in the DNA sequence of an organism.
The size of a mutation can range from one single nucleotide to an entire region in a chromosome. There are many types of mutations (Figure 1). For example, a point mutation occurs when a single nucleotide is replaced with another single nucleotide. An insertion mutation occurs when an extra piece of DNA is added to a chromosomal region. A deletion mutation, conversely, occurs when a part of the DNA sequence is missing from the genome.
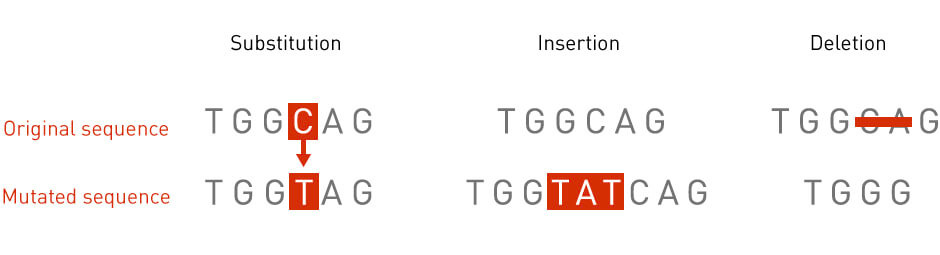
Figure 1: Types of mutations. Diagrams illustrating three types of mutations: point mutation, deletion, and insertion.
Consequences of mutations
The consequence of having a mutation in the genome also varies. A mutation can have no effect on an organism if the mutation occurs in a region that does not affect gene products or gene functions. This can happen if the mutation occurs at the non-coding region of the DNA or if the mutation occurs at a coding region, but does not change the final amino acid sequence of the gene product. This type of genetic alteration is called a silent mutation. A silent mutation is the major type of mutation which occurs in our genome.
If a mutation changes the amino acid sequence of a gene product or alters its expression, phenotypic consequences may occur, which could then affect the fitness level of the organism in its environment. A mutation is advantageous when its phenotypic consequence allows the organism to be more adaptable to the environment. Conversely, a mutation is deleterious when the organism becomes less adaptable to its environment.
Natural causes of mutations
Mutations can be accumulated naturally in two ways. It can either be passed down from parent to offspring, which is called a hereditary mutation or acquired throughout an organism’s lifetime. The latter, Acquired mutations, occur intrinsically from errors during DNA replication in actively dividing cells. They can also be induced extrinsically by external factors, such as UV radiation and free radicals.
Organisms acquire mutations through intrinsic and extrinsic factors regularly, which cause variations in our genome. Variation caused by genome mutation is essential for the long-term survival of species because it provides the basis for natural selection, evolution, and adaptation to the changing environment over time.
Laboratory methods to generate mutations
Mutations can also be intentionally introduced in a laboratory setting. Over the years, scientists have developed techniques to generate genetic mutations in model organisms. These techniques are powerful tools for scientists to study gene and protein functions.
Random mutagenesis
Random mutagenesis is a method that generates mutations in a random fashion (Figure 2). Random mutagenesis can be achieved by using external mutagens, such as radiation or mutagenic chemicals (1). It can also be achieved through the use of error-prone DNA polymerases that introduce wrong nucleotides during DNA replication (2). Random mutagenesis was one of the earliest tools available to introduce mutations in the genome and has continued to be utilized today to generate libraries of randomized gene and protein sequences.
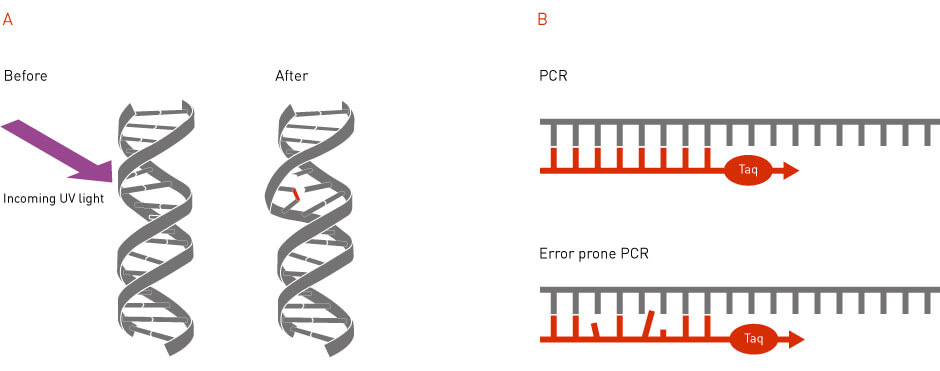
Figure 2: Random mutagenesis. In random mutagenesis, the mutations can be generated by physical or chemical mutagens or by the usage of error-prone DNA polymerases.
Site-directed mutagenesis
Site-directed mutagenesis is a method that generates specific nucleotide changes at a desired genome location (3). The technique is achieved through PCR reactions using primers that contain desired mutations. The mutant plasmid can then be obtained through sequential steps of PCR purification, template degradation, E. coli transformation, and plasmid isolation (Figure 3).
Site-directed mutagenesis is a powerful technique to introduce targeted mutations. As long as the primers are able to hybridize to the desired locus, more than one mutation can be introduced to the primers. Therefore, the mutations generated by site-directed mutagenesis can be single-point mutations, multiple-point mutations, insertions, or deletions. Therefore, site-directed mutagenesis is a versatile tool to introduce a wide range of mutations into the genome.
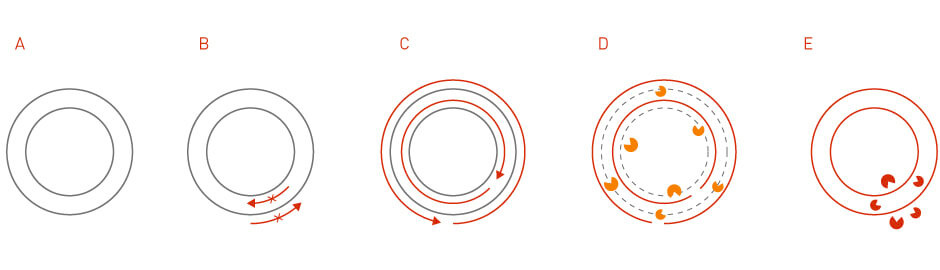
Figure 3: Site-directed mutagenesis. In site-directed mutagenesis, the specific mutation is introduced into the primers that are used for the initial PCR reaction.
Gene knockout and gene targeting by homologous recombination
Homologous recombination (HR) is a naturally occurring process that facilitates the exchange of genetic materials between two similar DNA sequences. It is involved in fixing deleterious DNA damage, such as DNA double-stranded breaks. It also promotes the exchange of genetic materials between homologous chromosomes in meiosis for spores and gamete production. Therefore, HR is a critical process that maintains genome integrity in a single organism, while promoting genetic variation in a population. Not surprisingly, HR is conserved in all domains of life.
The conservation of HR throughout evolution has allowed scientists to perform high-precision gene knockout and gene targeting in many model organisms, including bacteria, yeast and mouse (4). In a typical gene-targeting scenario, a host gene is replaced with a selectable marker. The selectable marker is PCR-amplified so that the flanking sequences are identical to the up-stream and down-stream sequences of the host gene. The flanking sequences are required for HR to guide and integrate the marker into the host genome at the desired locus (Figure 4).
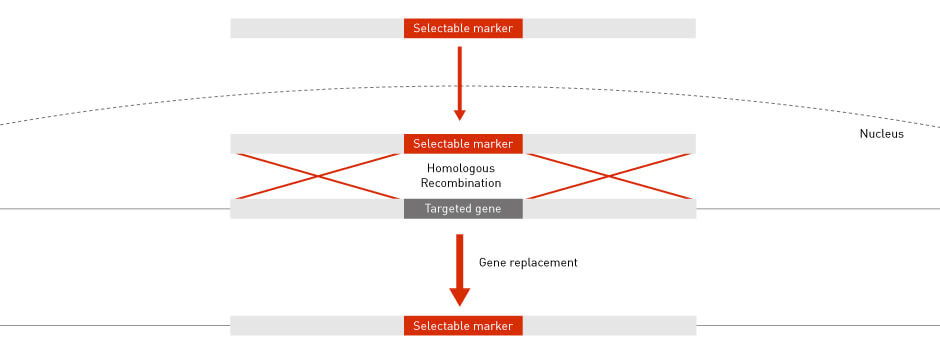
Figure 4: Gene targeting by HR Homologous recombination permits the specific targeting and knocking out of endogenous DNA sequences.
Deletion mutation collections
Using the HR-based gene knockout approach, scientists have systematically knocked out every gene in the entire yeast genome, creating a yeast deletion library of ~5000 single-gene deletion mutants. Since then many more library collections have become available. Some of them add epitope tags to host genes, while others are made in other organisms.
With the help of high-precision robotics, the availability of deletion libraries has permitted scientists to perform high-throughput screening assays to probe for drug targets, identify genetic interactions on a genome-wide level, and study the complexity of genetic networks.
For more information on high-throughput screens and screening robots, please click here.
Reference
Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989 Apr;77(1):51–9.

