Bacteria-genetic-screening
Bacteria-genetic-screening – E. coli Synthetic Genetic Array (eSGA)
Jay Yang, Singer Instruments, Roadwater, TA23 0RE
Summary
E. coli Synthetic Genetic Array (eSGA) analysis involves the manipulation of high-density bacteria arrays to monitor bacterial genetic interactions. In a typical eSGA screen, a query strain is systematically conjugated to a genome-wide arrayed collection of single gene mutants. After rounds of robotic pinning and duel marker selection, E. coli strains harboring both gene mutations can be assayed for growth defects. Systematic and quantitative identification of genetic interactions allows the construction of large genetic networks, revealing functional dependency and pathway redundancy in E. coli.
Principles of ESGA
The generation of double mutants in eSGA relies on the principle of conjugation, which is the horizontal transfer of genes between bacteria [1].
In conjugation, a high frequency of recombination (Hfr) donor strain that contains the integrated fertility (F) factor can transfer genetic materials to an F– recipient strain in a unidirectional manner. Once inside the recipient strain, the donor DNA can be integrated into the recipient genome by homologous recombination [2].
Based on this principle, eSGA is developed using the following steps [3]
- Donor mutant is constructed in the E. coli Hfr strain by replacing the gene of interest with a chloramphenicol (Cm)-resistance marker.
- The recipient mutant arrays are constructed in the E. coli F– strain containing ~4000 single gene deletion mutants marked with a kanamycin (Kan)-resistance cassette.
- After mutant construction, the donor mutant is pinned onto the recipient mutants on LB plates to allow the transfer of query DNA to the donor chromosome via conjugation.
- The double mutants are then selected on LB plates that contain both Cm and Kan. The resulting colonies are then digitally imaged and analyzed for growth fitness.
Theory Behind Genetic Interactions
If two genes were functionally unrelated, the double mutant containing these two gene mutations would exhibit a phenotype that is approximately the product of the phenotypes of the individual mutants.
However, when mutations in two gene products produce a phenotype that deviates from each gene’s individual effect, these two genes exhibit a genetic interaction. There are two general categories of genetic interactions: aggravating interactions and alleviating interactions (Figure 2A).
An aggravating interaction occurs when a double mutant exhibits a phenotype that is more severe than expected from the phenotypes of individual mutants. This type of interaction indicates that the gene products function in redundant parallel pathways, and highlights the robustness of the molecular network in tolerating genetic variations [4].
An alleviating interaction occurs when a double mutant exhibits a phenotype that is less severe than expected from the phenotypes of individual mutants. This type of interaction indicates that the gene products operate in concert or in series within the same pathway [4].
Genes that function in the same pathway tend to have similar genetic interaction profiles. Therefore, identifying genetic interactions can reveal functional relationships between genes and biological pathways [4].
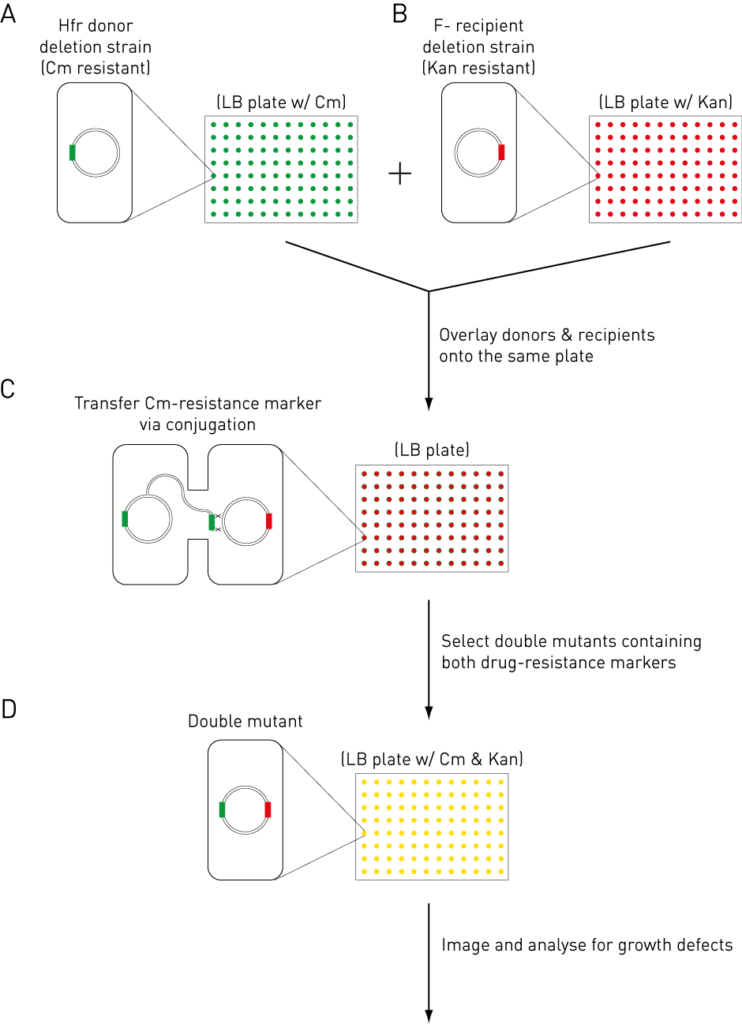
Figure 1: eSGA Principles and Steps.
Aggravating interactions are shown in red, while alleviating interactions are shown in green. See text for details.
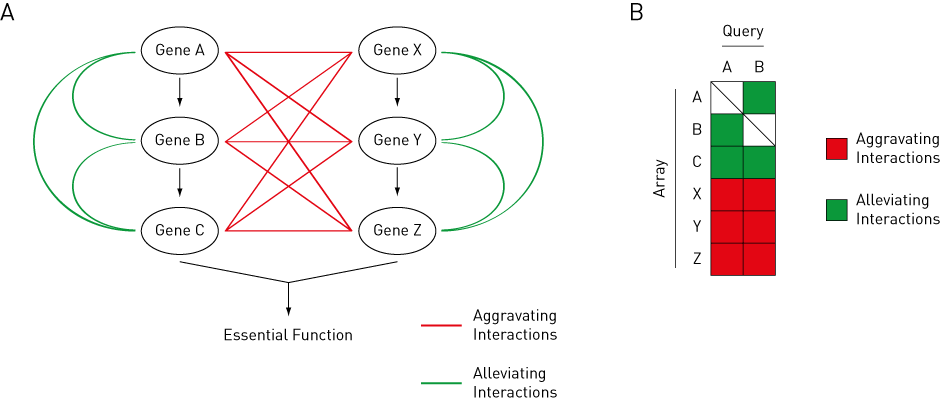
Figure 2: Theory Behind Genetic Interactions.
Aggravating interactions are shown in red, while alleviating interactions are shown in green.
(A) Pathway ABC and XYZ both contribute to an essential function in a cell. Gene A shows aggravating interactions with Gene X, Y, and Z, but shows alleviating interactions with Gene B and C.
(B) Gene A and B show similar genetic interaction profiles to each other.
Building Genetic Interactions into Biological Networks
Genes in the same pathway often display aggravating interactions with genes in parallel and functionally redundant pathways. On the other hand, genes often display alleviating interactions with one another when their products function in the same pathway or protein complex [5]. Based on these two principles, constructing two-dimensional clusters of all pair-wise genetic interactions could reveal the roles of individual genes in various cellular processes and identify components of a protein complex, or those within a specific pathway [4].
Applying ESGA to Other Bacterial Systems
Because conjugation is a widespread phenomenon in prokaryotes [1], the ESGA technique can be applied to study genetic interactions in other bacterial systems. Indeed, E. coli has been shown to efficiently transfer non-replicating plasmids to other bacterial species [6]. Therefore, with various recipient gene deletion mutant collections being available [7,8], it is possible to apply eSGA to systematically analyze pair-wise genetic interactions in other prokaryotes.
Working With Libraries
High-throughput eSGA screening can be done quickly and easily using automated robotics, such as the Singer ROTOR+, which can rapidly pin high-density arrays of bacteria and facilitate strain conjugation and selection. In addition, the ROTOR+ screening robot provides an automated platform to simultaneously produce multiple experimental replicates.
Bacteria Genetic Screening Applications
High-throughput, quantitative analyses of genetic interactions in E. coli
Abstract Typas A, Nichols RJ, Siegele DA, Shales M, Collins SR, Lim B, Braberg H, Yamamoto N, Takeuchi R, Wanner BL, Mori H, Weissman JS, Krogan NJ, Gross CA
Large-scale genetic interaction studies provide the basis for defining gene function and pathway architecture. Recent advances in the ability to generate double mutants en masse in Saccharomyces cerevisiae have dramatically accelerated the acquisition of genetic interaction information and the biological inferences that follow. Here we describe a method based on F factor-driven conjugation, which allows for high-throughput generation of double mutants in Escherichia coli. This method termed genetic interaction analysis technology for E. coli (GIANT-coli), permits us to systematically generate and array double-mutant cells on solid media in high-density arrays. We show that colony size provides a robust and quantitative output of cellular fitness and that GIANT-coli can recapitulate known synthetic interactions and identify previously unidentified negative (synthetic sickness or lethality) and positive (suppressive or epistatic) relationships. Finally, we describe a complementary strategy for genome-wide suppressor-mutant identification. Together, these methods permit rapid, large-scale genetic interaction studies in E. coli.
References
Looking for screening tools to accelerate your research?
ROTOR+: the only microbial screening solution capable
of pinning a million colonies an hour.