Case Study: High-throughput microscopy of arrayed strains with strain library imaging protocol: SLIPping into awesome microscopy in the Huang Lab at Stanford University
Amanda Miguel¹, Handuo Shi¹, Nripesh Dhungel², KC Huang¹
¹Stanford University, Stanford, CA 94305, USA.
²Singer Instruments, 1250 45th St. #330, Emeryville, CA 94608, USA.
What is it?
SLIP (Strain Library Imaging Protocol) is a novel methodology developed by the KC Huang lab at Stanford University to rapidly image hundreds of thousands of arrayed bacterial cells in one microscopy session.
What Were the Limitations of Microscopic Imaging of Multiple Bacterial Strains at once?
Normally, imaging of bacteria is limited by three things: 1) the size of the surface the cells are placed on, which physically limits the number of strains (or types of organisms) that can be imaged simultaneously; 2) the spacing between the strains you image, since sufficient spacing is needed to reliably distinguish between one strain and the next without fear of mixing; 3) capacity for human error, which contributes to inconsistency and failure. SLIP solves these problems by bridging the microscope and the physical limitations of imaging using pre-existing common lab tools and a clever application of software.
What is the Solution that Slip provides?
Large repositories of strains such as bacteria are often stored as libraries in arrays, meaning they are frozen down and stored in plastic plates that contain wells devoted to individual strains. These arrayed plates, while varying in the number of wells, are all of a standard size. Recognizing this, we developed SLIP around the idea that if one can construct a surface the same size as these storage plates, an entire plate with arrayed strains can be imaged in a single microscopy session. This protocol allows us to image from 96 to as many as 1536 different populations on the same surface.
To image cells, we simply use the RePads created by Singer Instruments, which were initially designed to manipulate libraries and allow transfer of colonies between agar plates or various different media, to move populations of cells onto an imaging pad. Notably, using this format gives us two important advantages: 1) because each well in an arrayed collection is a fixed distance from every other well, the separation between strains is guaranteed to be fixed; and 2) because of the fixed spacing, knowing the position of one well enables the computer to extrapolate the positions of all other wells. This means that if the position of the first well is known, the microscope can automatically image the entire plate in a fast and robust manner. This procedure is achieved by the SLIP software, and coupled with an electrical triggering strategy to reduce the communication time between the microscope and computer by ~90%, it is now possible to image ~100 strains in as little as 5 minutes.
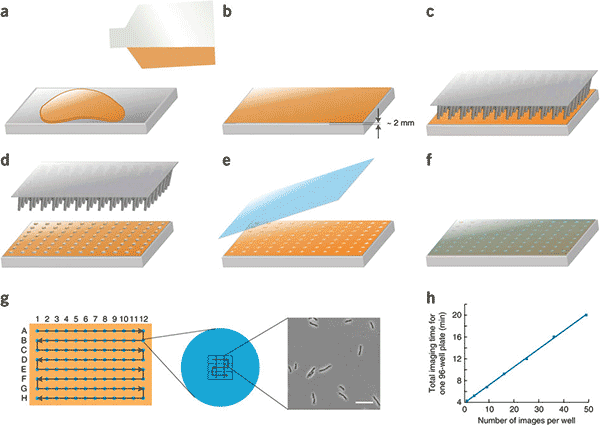
Figure 1: SLIP imaging protocol
Conclusion and the Future!
SLIP is a powerful technology that allows for the high-throughput imaging of large arrayed libraries of strains. While the Huang lab focuses primarily on bacterial collections and arrays, the technology is flexible enough to be extended to other organisms of different scales with relative ease. We are currently exploring ways to expand this technology, such as through high-throughput single-cell time-lapse imaging and multi-channel fluorescence imaging. We are also working with Singer Instruments to determine ways to improve SLIP’s automation, making it easier than ever to use SLIP for diverse projects. Our immediate goal is to utilize Singer Instrument’s ROTOR+ high-density arraying robot to automate the pinning and preparation of strains for SLIP. Stay tuned!