High-throughput microfluidic loading of microbial libraries
Nicholas Csicsery, Elizabeth Stasiowski, Garrett Graham, Gregoire Thouvenin,
William Mather, Michael Ferry, Scott Cookson and Jeff Hasty. University of California, San Diego, La Jolla, CA, USA.
Summary
Laboratory automation has accelerated the rate at which microbial strain libraries can be generated. These libraries include those which study the effects of gene knockouts [1] fluorescently labeled genes [2], or novel synthetic constructs [3]. However, screening tools are often limited to batch systems.
Towards this end, multiplexed microfluidic platforms serve as a means to survey libraries of strains in parallel under continuous growth conditions. Existing multiplexed devices [4] rely on growth in liquid media before loading onto devices. With the ROTOR+ from Singer Instruments, it is now possible to simultaneously load a SBS-formatted microfluidic device directly from an agar plate [5]. This enables the real-time dynamic interrogation of microbial strain libraries, a useful tool for the study of biological networks and screening of gene circuits in synthetic biology.
Methods
A multiplexed microfluidic array is created by spacing 2,176 strain banks at a 6,144 SBS density, or 1.125 mm apart. Strain banks are separated into two 17×64 sections and fed media through a manifold-based fluidic assembly. Cells are spotted into a 500-um diameter circular spotting region, from which they are wetted and grow towards the media channel interface. At this interface, exponentially growing cells are imaged with outgrowing cells washed away towards the outlet channels.
In preparation for loading onto microfluidic devices, strain libraries are grown on agar-based media in PlusPlates from Singer Instruments. Libraries, initially stored and grown at a 96 or 384 SBS density, are arrayed onto four fresh agar plates at a 1,536 SBS-density using a Stinger attachment for the ROTOR+ or a PIXL from Singer Instruments. The four plates are finally combined into an 6,144 density arrangement mirroring that of the microfluidic array before direct transfer onto the Polydimethylsiloxane (PDMS) component of the microfluidic assembly (Fig.1).
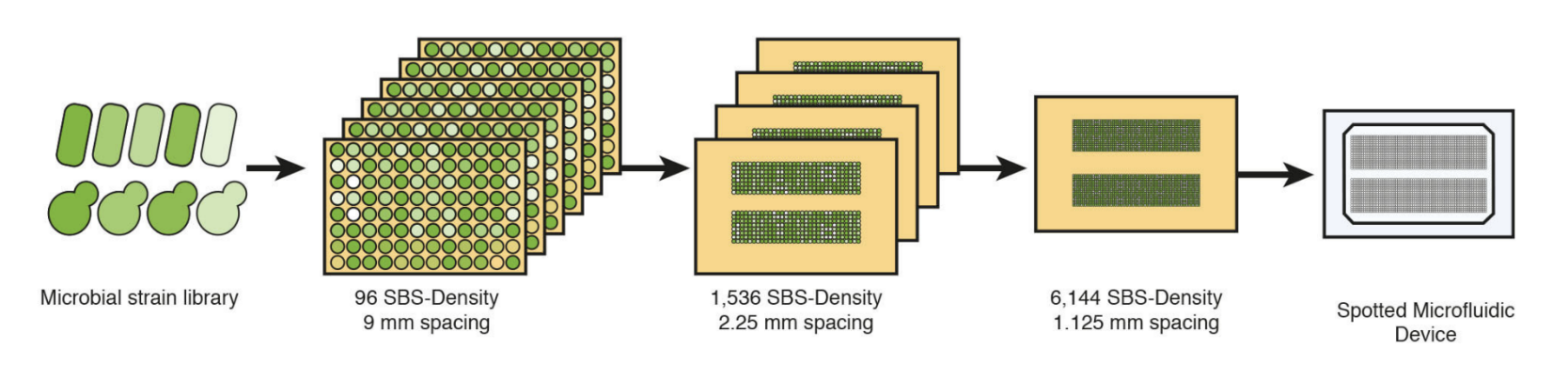
Figure 1: Microfluidic loading with the ROTOR+ from Singer Instruments. Strain libraries of E. coli or S.cerevisiae are grown on 96 SBS-density PlusPlates, and re-arrayed at an eventual 6,144 SBS-density before colony deposition onto microfluidic components.
To accurately transfer microbial colonies from agar plates to PDMS, cells are initially spotted as alignment markers onto an acrylic microfluidic-to-ROTOR+ adapter. The PDMS device is then aligned to these alignment markers, feature-side-up, where surface adhesion prevents slipping and misalignment of the PDMS. The PDMS-acrylic assembly and glass slide are exposed to oxygen plasma, after which colonies are transferred from the 6,144 density agar plate to the PDMS device using a standard ROTOR+ from Singer Instruments 6,144 short RePad. PDMS is bonded at 37°C for 1-2 hours before wetting with the respective growth medium (Fig.2).
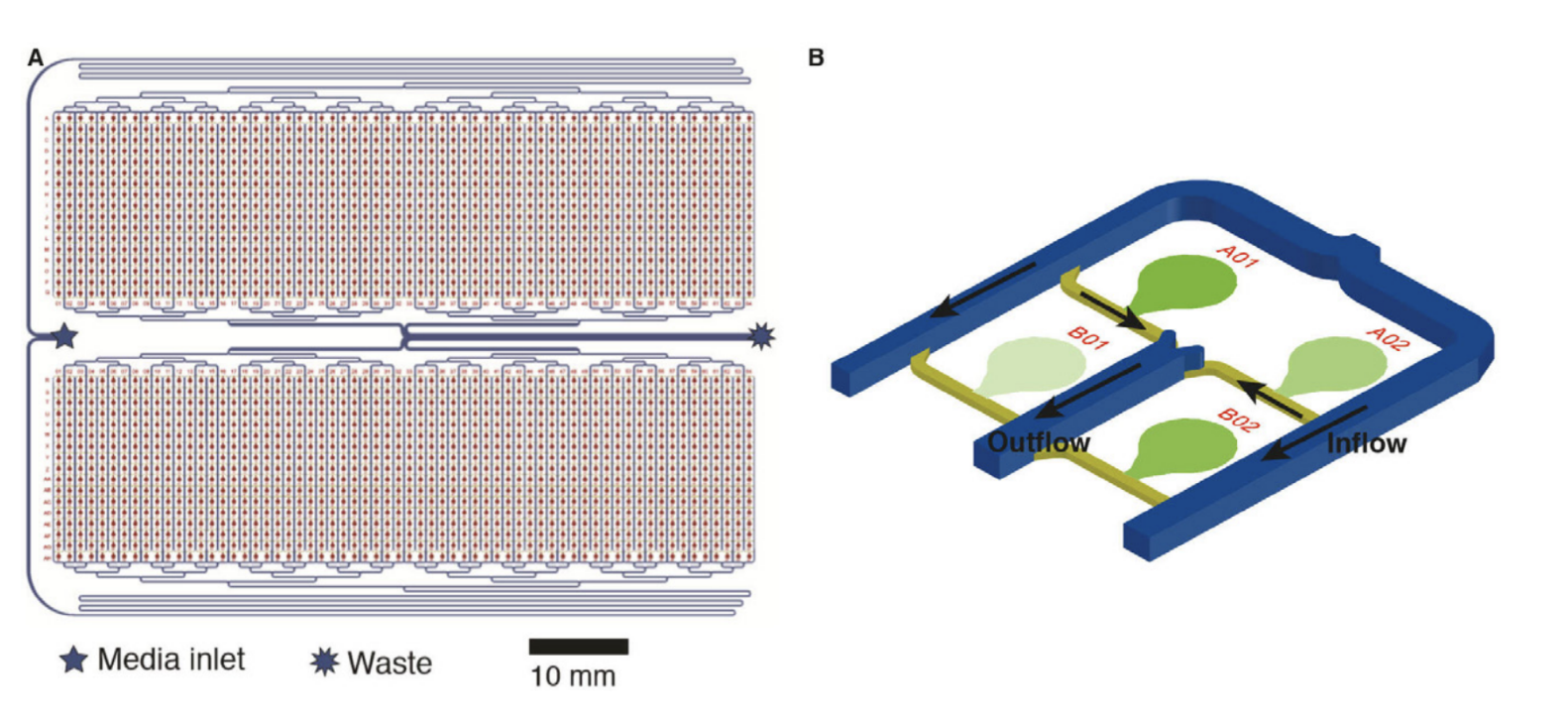
Figure 2: A 2,176-strain microfluidic device (A) Something. (B) A subsection of 4 of the 2,176 strain banks on a microfluidic device. Cell spotting and growth regions are highlighted in green. Inflow and outflow channels are marked with directional arrows.
The loading of these highly multiplexed microfluidic devices with the ROTOR+ from Singer Instruments was validated using previously engineered fluorescent libraries of both E. coli and S. Cerevisiae [3,2]. Libraries were assessed for viability after spotting, cell retention, and measurable fluorescent signal. After verification of successful growth and imaging of these strains, the libraries were subjected to temporal environmental perturbations, including carbon-source shifts, hyperosmotic stress, and exposure to numerous inducing compounds (Fig.3).
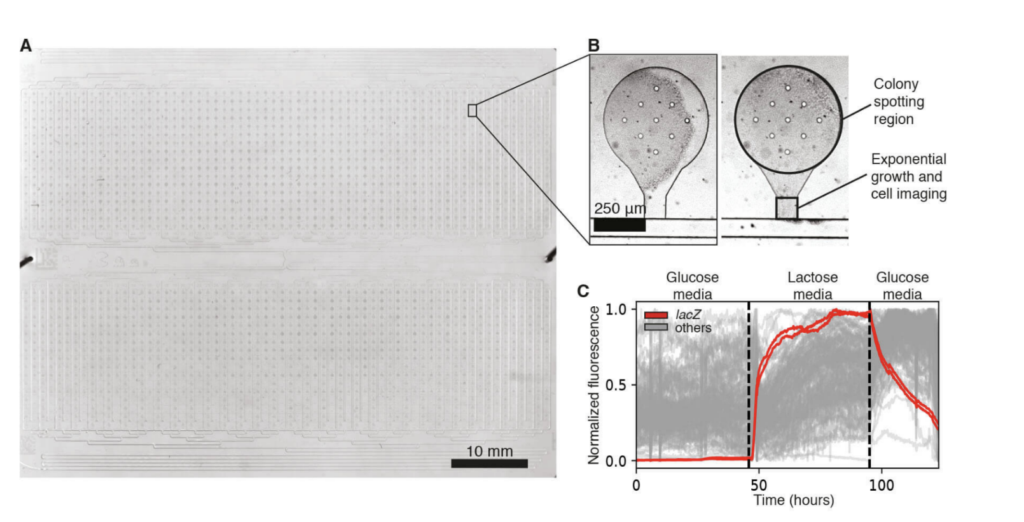
Figure 3: An E. coli strain library on a 2,176-strain microfluidic device. (A) A single transmitted light image captures the full device. (B) A microfluidic strain bank after initial spotting and chip wetting (left) and after 16 hours of growth (right). (C) 50 fluorescence traces were obtained from an E. coli promoter library, for which a different promoter drives the expression of GFP in each strain, during a glucose-lactose-gluce carbon source shift. Two lacZ promoter strains are highlighted in red, while the traces from 48 other promoters are shown in gray.
References
3. Alon Zaslaver, Anat Bren, Michal Ronen, Shalev Itzkovitz, Ilya Kikoin, Seagull Shavit, Wolfram Liebermeister, Michael G. Surette, and Uri Alon. A comprehensive library
of fluorescent transcriptional reporters for Escherichia coli. Nature Methods, 3(8):623-628, 2006.
5. Stasiowski E, O’Laughlin R, Holness S, Csicsery N, Hasty J, Hao N. A Microfluidic Platform for Screening Gene Expression Dynamics across Yeast Strain Libraries. Bio Protoc. 2023 Nov 20;13(22):e4883. doi: 10.21769/BioProtoc.4883. PMID: 38023791; PMCID: PMC10665637.

Jeff Hasty
Professor, BENG
Professor, Department of Molecular Biology,
School of Biological Sciences
Check out this 2023 Bio-protocol by Jeff and colleagues for more details on the use of ROTOR+ to support screening of gene expression dynamics [5].
Scale up your workflow with the ultimate high-throughput screening combo
Combining ROTOR with PIXL lets you pick and re-plate
at densities up to 6144 and at a rate of nearly a million colonies an hour.