Using ROTOR+ to investigate viral-host protein interactions using a selective ablation assay
A SARS-CoV2 case study
In this article, Professor Rodney Rothstein details the novel approach his lab is taking to study viral-host protein interactions using a selective ablation assay and ROTOR+ to achieve high-throughput genome-wide screens within 6 days.

Background
COVID-19, the respiratory syndrome caused by infection with the SARS-CoV-2 coronavirus has resulted in greater than 1.2 million deaths worldwide causing a strain on medical care facilities in many countries and a significant economic impact worldwide [1-3]. The SARS-CoV-2 virus is a zoonotic coronavirus that crosses from bats to humans in a form that could spread rapidly via respiratory droplets and aerosols. Two other major respiratory syndromes caused by coronaviruses, SARS and MERS, have emerged within the last two decades [4, 5]. Thus novel coronavirus infections are likely to be a continuing threat to human health.
Coronaviruses are enveloped positive-strand RNA viruses in the order Nidovirales. All coronaviruses contain a spike protein that spans the envelope membrane, it is important for host receptor binding and infection, and gives these viruses their crown-like appearance in electron micrographs [6]. Coronaviruses contain the largest genomes of any positive-strand RNA viruses – about 30 Kb – in part due to encoding an RNA exonuclease that acts as a proofreading mechanism during genome replication [7]. The first two-thirds of the viral genome is translated into two large polyproteins that are processed into individual non-structural proteins through the action of viral-encoded proteases. These non-structural proteins rearrange membrane structures in the host and form the viral replicase. The latter third of the genome encodes the structural proteins nucleocapsid (N), spike (S), an envelope protein (E) and M protein as well as several open reading frames that vary widely among coronaviruses [6].
Although much has been learned about the SARS-CoV-2 virus through intensive study, much more needs to be understood to combat this virus as well as likely novel emerging infectious coronaviruses. Yeast can serve as a flexible experimental platform to help study SARS-CoV-2 safely. Although positive-strand RNA viruses have not been found in yeast, several positive-strand RNA viral replication systems have been constructed by expressing the viral RNAs from yeast promoters [8-10]. Individual viral proteins from SARS-CoV-2 have also been expressed in yeast to identify genetic interactions that are common to eukaryotes [11]. In addition, viral proteins have been expressed in yeast as a way of identifying enzymatic activities of viral proteins as well as to evaluate inhibitors of those activities [12].
Our approach
We are using the Singer Instruments ROTOR+ to screen the yeast gene disruption library for virus-host genetic interactions. Our method, Selective Ploidy Ablation (SPA), a type of selective ablation assay, provides a high throughput, mating-based approach to move expression plasmids into an arrayed yeast library [13]. The method relies on a donor strain in which all sixteen yeast chromosomes contain a centromere-proximal counter-selectable marker (URA3) and the galactose-inducible promoter from the GAL1 gene (Figure 1). Growth on galactose induces expression and disrupts centromere function in these strains leading to chromosome loss during mitotic growth. In a haploid context, this strain dies when grown on galactose. In a diploid heterozygous for the CEN-marked chromosomes, growth on galactose leads to specific loss of the CEN-marked chromosomes and all the accompanying genetic information. Thus a diploid heterozygous for all sixteen chromosomes can become a haploid simply by mitotic growth on galactose.
With the Singer Instruments ROTOR+, the SPA procedure can be used to perform plasmid transfer into a library of strains. The SPA donor strain is first transformed with an expression plasmid containing the viral gene of interest using st as the selectable marker. Transformant cultures are grown in dropout medium overnight and then spread onto YPD medium in Singer PlusPlates™ to form a confluent lawn. A compatible yeast library of opposite mating type is freshly pinned onto YPD and both sets of plates are grown overnight. The following day, the donor lawn and the library array are pinned together for mating, grown for 6-8 hours on YPD to allow mating, and then pinned to -LEU dropout medium containing galactose as a carbon source. Plates are incubated at 30°C for two days on galactose to destabilize the donor strain chromosomes, then pinned to -LEU dropout containing galactose and 5-fluoro-orotic acid (5-FOA) to counter-select any remaining strains that still contain donor chromosomes. Plates are incubated 2-3 days on 5-FOA for haploid growth before imaging. Colonies containing viral expression plasmids are compared to an empty vector control to determine fitness effects in a library of yeast mutants.
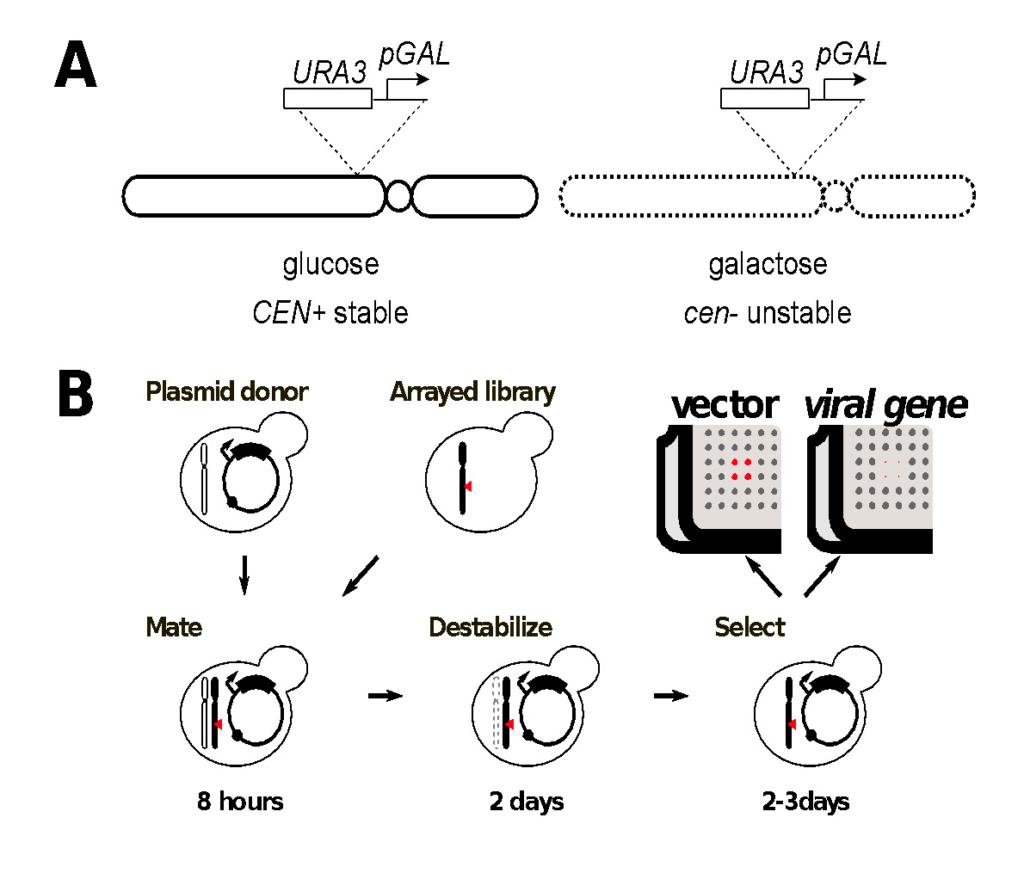
Figure1. Genome-wide screens of the yeast gene disruption library by selective ploidy ablation (SPA).
A. Deception of yeast chromosomes contains an integration of the GAL1 promoter and a URA3 marker proximal to its conserved centromere sequence. Activation of transcription from the GAL1 promoter inactivates the centromere landing to its destabilization during mitotic growth.
B. Plasmid transfer from the universal donor to a recipient strain or library of strains is achieved by mating the plasmid-containing universal donor strain to the recipient for 8 hours. Strains are then transferred to glactose-containing dropout medium for 2 days to select for the plasmid and destabilisation of the donor chromosomes. Strains are then transferred to 5-FOA dropout medium for 2-3 days to counter select any remaining donor chromosomes, while mainitng the selection for the transferred plasmid. Negative synthetic genetic interactions are identified by colonies that grow with the empty vector control (red in the cartoon) but fail to grow when a viral gene is expressed.
In summary
The SPA screening procedure is efficient due to the mating-based transfer, and rapid because it does not require sporulation and haploid selection. The genome-wide screens can be accomplished in six days using ROTOR+. Imaging and colony size measurement is performed using an R package version (https://github.com/EricEdwardBryant/screenmill). This package provides R functions for processing multiple types of plate images to generate colony size measurements. Notably, the package also allows for the processing of time-series images suitable for visualizing growth curves (Figure 2).
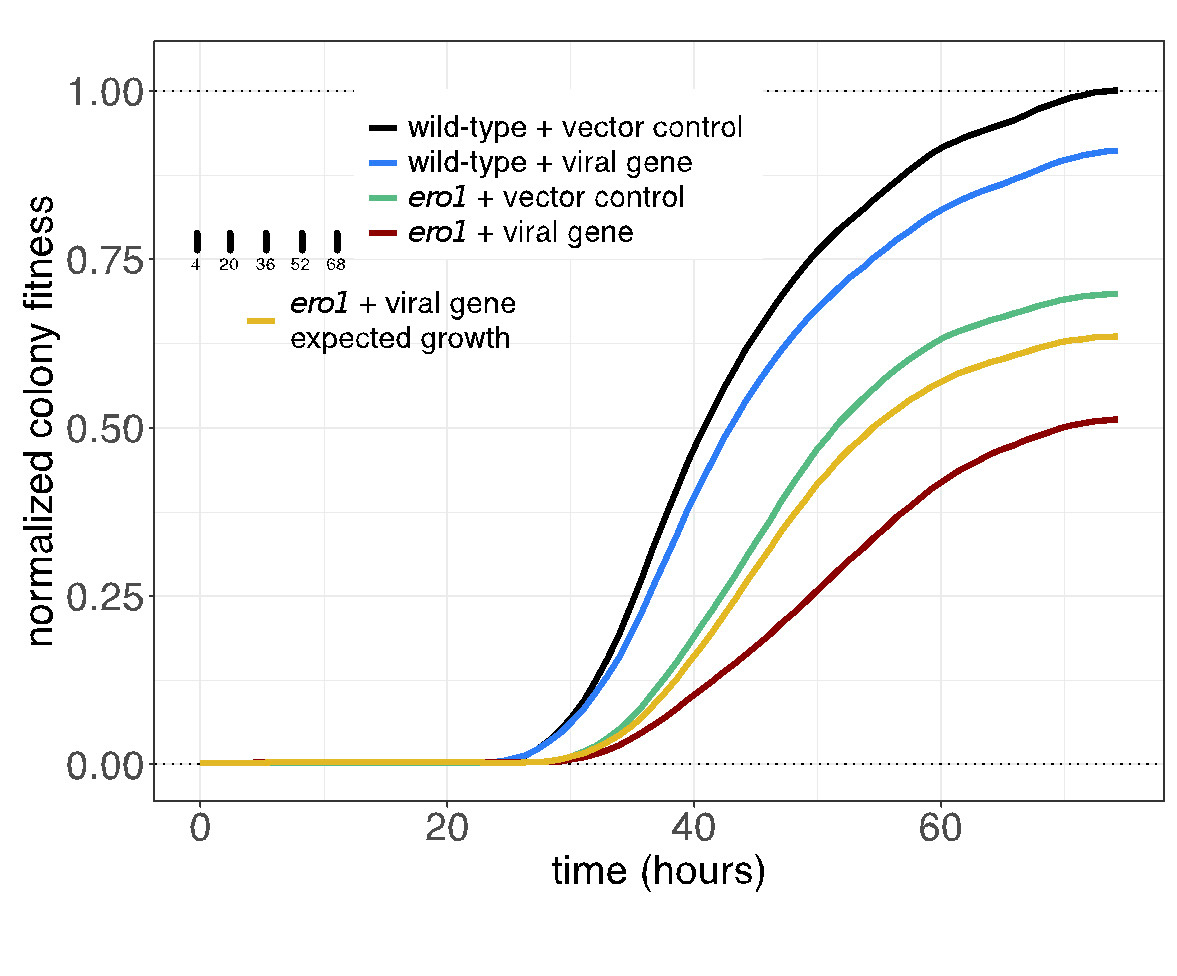
Figure2. Yeast cultures expressing the empty vector or the viral gene clone are equalized by 0D(600) and spotted onto Singer PlusPlates.
The plates are incubated and are scanned once an hour over three days to monitor growth. Image density at each colony position is measured using the sceenMill R package. Growth curves are plotted from measurements of colony density (selected colony timepoint in inset). Confidence intervals of mean growth values are indicated by shading. All values are normalized to maximal growth wild-type strain containing the empty vector (black line). Exected growth of the viral genes in the ero1 strain (yellow line) is calculated from the product of the wild-type strain expressing the viral gene (blue) and the mutant strain expressing the vector control (green line) at each time point. The observed growth of the ero1 mutant containing the viral gene (red line) is less than expected growth. Indicated a negative genetic interaction. Since the confidence intervals of these curves do not overlap, the difference is significant.
References
Looking for an automation solution with
the throughput for large-scale genetic screens?
With ROTOR+ you can screen the whole yeast gene disruption
library for SARS-CoV-2 virus-host genetic interactions in under 6 days.

