Dr Michal Breker on Chlamydomonas automation
Industrial solutions from academic research
Fiona Kemm1 and Dr Michal Breker-Dekel2
1 Singer Instruments, 2 The Hebrew University of Jerusalem

Introduction to Chlamydomonas research
Dr Michal Breker-Dekel has risen to become a leading light in efforts to cement Chlamydomonas reinhardtii as the preferred model organism for the plant superkingdom. Work she undertook as a postdoc with Prof. Fred Cross at the Rockerfeller Institute, “to establish high-throughput automated loss-of-function genetic screens of Chlamydomonas essential genes for the first time”, has built the foundations from which concerted efforts to study plants through Chlamydomonas could begin. Now the PI of her own lab at the Hebrew University of Jerusalem, Michal’s focus is on understanding chloroplast biology and its crucial ties with carbon fixation from metabolism: “It’s like the essence of our life on earth and the energy for the vast majority of organisms,” she says.
Establishing a high-throughput platform in Chlamydomonas
Michal first encountered ROTOR while working towards a PhD in yeast genetics in the lab of Prof. Maya Schuldiner at Israel’s Weizmann Institute of Science. It was from here she transitioned to her postdoc with Professor Fred Cross, where her escapades with ROTOR+ turned from yeast to Chlamydomonas.
Using the robot, Michal was able to isolate 10,000 temperature sensitive lethal mutants and characterize 340 cell cycle mutants (~170 previously unidentified) with near comprehensive representation of all essential cell cycle functional modules. She further published her high-throughput experimental pipeline, demonstrating how a temperature sensitive mutation library can be reproduced in any organism cultivatable on agar, highlighting ROTOR+ as “very important for efficiency in this procedure,” (Breker et. al., JOVE 2016; Breker et. al., Plant cell 2018). Still relying on ROTOR+, Michal reflects that “The design of ROTOR+ is so good, it’s worked robustly for so many years.”

Photo credit: Noemie Alon.
Saving the world through plants
Towards the end of 2019, Michal started her own lab at the Hebrew University of Jerusalem. Juggling the challenges of being a new mother, a new PI, the onset of COVID-19 and growing political unrest, Michal set up her high-throughput automation lab focused on understanding the systems biology of the Chlamydomonas chloroplast. “These are fundamental organelles for our lives on earth, central hubs for carbon fixation and metabolism.” describes Michal. “Understanding the fundamental science behind this can therefore pave the way for new sustainable technologies.”
With a great deal of this fundamental research under her belt, Michal says it’s now her “personal mission to take research beyond the academic setting and translate it into applicative solutions that can help humanity.”
And to do that she knew exactly which robot she wanted by her side: “When I set up on my own there was no question that I wanted the ROTOR+ in my lab. I think it’s such an excellent robot. I don’t think there’s any other product quite like it. It allows us to do so many experiments and it just works all the time.”
Photo credit: Noemie Alon
Michal later added a PIXL colony picker and a Sporeplay+ to the suite, creating her very own high-throughput powerhouse for genetic screening and linkage analysis in Chlamydomonas. But she’s not stopping there:
“We’re considering maybe integrating our PIXL with hotels and a robotic arm to feed the plates in and out. We work on generating very large collections of mutants and it means someone standing and doing manual loading all day.”
High-Throughput automated Chlamydomonas experiments
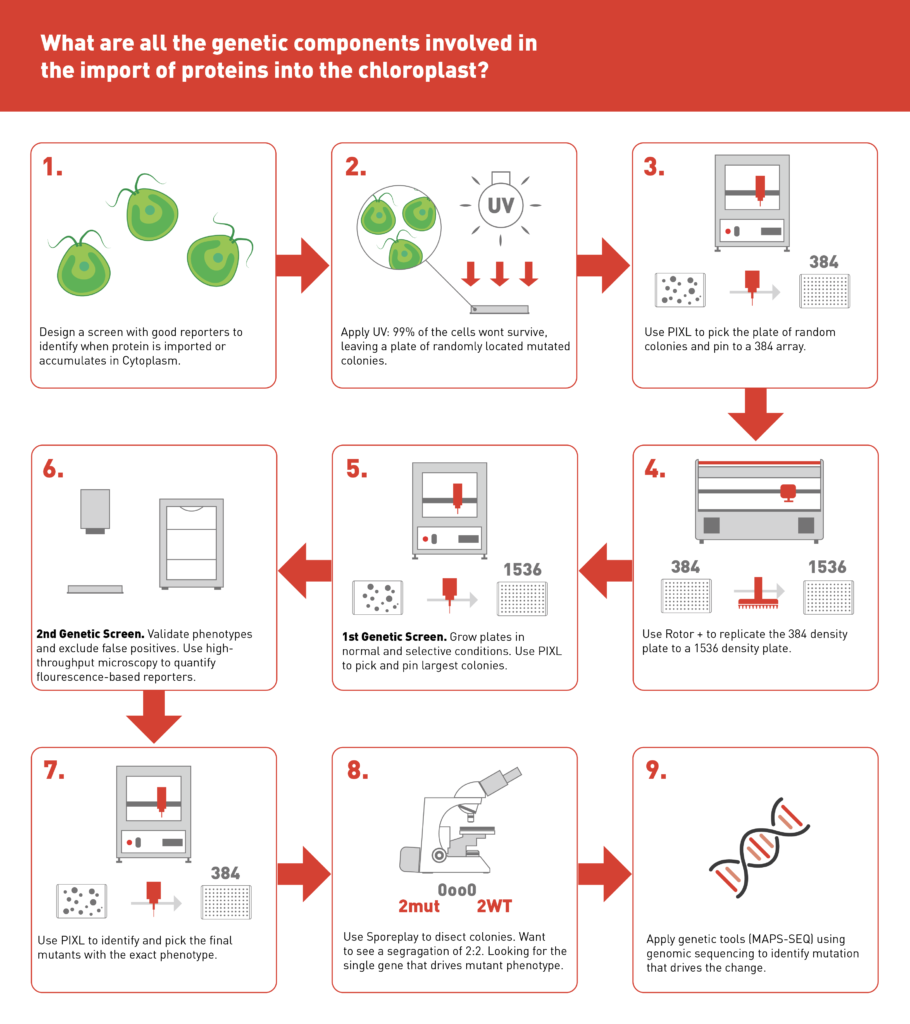
Since Michal’s initial robotics pipeline was first published in 2016, she has developed and improved on it. Michal now uses PIXL, ROTOR+ and SporePlay+ to create, maintain and investigate Chlamydomonas reinhardtii libraries.
“We build mutant collections up to 200 000 strong to reach good genetic saturation. We pick them with PIXL and maintain them with ROTOR+”

Dr Michal Breker
The Breker Lab, Hebrew University of Jerusalem

Photo credit: Noemie Alon
The classic pairing of PIXL and ROTOR+ efficiently picks the randomly mutagenised Chlamydomonas and replica-plates it in an arrayed format for temperature sensitivity assays (Breker et. al., JOVE 2016). Once the mutants are identified, SporePlay+ is essential to narrowing down the target pool through tetrad dissection as the first genetic analysis step. This robotic pipeline allows the Breker lab and their collaborators access to a huge library of temperature sensitive mutants linked to nearly all of the essential pathways in Chlamydomonas.
Results from Chlamydomonas automation
By using high-throughput automation Michal has already achieved some great things:
- Michal’s genetic screens of the fundamental biology of Chlamydomonas chloroplasts advance the identification of genes essential to multiple cell pathways; the current focus is chloroplast biogenesis and protein quality control. Chloroplasts and essential genes are both highly conserved between Chlamydomonas and the plant kingdom. “Any genetic component we find that’s important to the import of proteins and efficient chloroplast function are genes to be targeted for in plants. All this requires our experience with unique genetic tools, screens and so much more.” This means that Michal can investigate plant resilience to stressful environmental factors, such as temperature range and light intensity.
- The Breker lab is collaborating with the Finkel lab to investigate the microbial ecosystem of plants. Together they are connecting genotype to phenotype, and back again. Together they co-culture environmentally isolated bacterial communities with the Chlamydomonas mutant collection. Through her automation, Michal has developed a reputation for excellence in Chlamydomonas. She collaborates with multiple labs to unblock their research using high-throughput genetic screens and the mutant libraries.
- Last year Michal co-founded CultivO2. Harnessing Chlamydomonas to form a unique ecosystem with animal cells to cultivate meat at a lower cost. By culturing the two together the photosynthesis of Chlamydomonas complements the respiration of the animal cells in a symbiotic relationship. The improved efficiency of oxygen and nutrient exchange reduces the media costs and increases animal biomass yield. In the future this will reduce the costs of cultivated meat alternatives on the end consumer.
Reflecting on 5 years of the Breker Lab
Access to high-throughput automation to generate genetic screens places the Breker lab at a pivotal point in Chlamydomonas research. Michal is a respected advisor to both industrial and academic labs, nurturing essential cross-talk between the two. “I don’t have to choose between them.” Michal researches both the fundamental mechanistic biological problems that industry doesn’t have the time for and explores the applications that academia has limited infrastructure for. The Breker lab are now adapting their protocols to apply their genetic toolkit to marine microalgae species, exploring broader ecological questions at a larger scale.
Michal noted that “as a student I didn’t have the mental space to tackle all the questions my curiosity wanted to explore.” High-throughput automation improves the efficiency of lab research and increases the speed at which questions can be answered. Michal went on to describe that “as a PI I can ask all the questions and feel involved in so many different projects”. We look forward to following her science at conferences and in the 5 publications planned for 2025.

Fiona Kemm | Research Scientist
Fiona plays a key role in ensuring our innovations perform as intended, while providing exceptional support to users. Her efforts in the lab contribute to the reliability of our robots, developing protocols and assays that demonstrate their unique capabilities. She has a BSc in Biochemistry, an MRes in Molecular Microbiology and a talent for BioArt.