Harnessing Natural Genetic Variation
High Throughput Automated Screening of the wild yeast microbiome.
Fiona Kemm1 and Dr Gianni Liti2
1 Singer Instruments, 2 Institute for Research on Cancer and Aging, Nice
The Liti Lab
Dr Gianni Liti is on a mission to understand genetic variation in his fantastic yeasts, but where to find them? The microbiome of oak trees in European and North African temperate forests, of course!
Gianni’s primary aim is to connect genetic variation in natural isolates of Saccharomyces yeasts to phenotypic variation. The Liti lab then uses this data to understand the cell’s complex biology, including traits relevant to cancer and aging in humans. This kind of research requires gathering and managing colossal numbers of colonies. But by using high-throughput automation Gianni can tackle his questions with ease.
Gianni Liti | Impulscience 2024
Credit: Alexandre Darmon, Art in Research for the Foundation Bettencourt Schueller
Why bioprospect the microbiome?
Over the last few thousand years humans have domesticated yeasts by creating environments with strong selection pressures. Through our collective love of all things alcoholic and fermented, humans sped up yeast evolution drastically. During his Post-doc with Professor Ed Louis, Gianni got his first taste of ROTOR. In collaboration with Jonas Warringer, they showed that domesticated yeasts have far less genetic differentiation than wild yeasts (Liti et. al., 2009, Nature).
Domesticated yeasts are also far more abundant in research because they are significantly easier to isolate than wild yeasts and so are disproportionately represented in the data. In fact, these wild yeasts can have as low as a 5% isolation success rate even on substrates, such as oak trees, where they are commonly found. By focussing on studying yeasts that, although more difficult to isolate, are more genetically diverse Gianni can better understand natural genetic variation. This increases the likelihood of his research unravelling the evolutionary histories of Saccharomyces species.

Credit: Alexandre Darmon, Art in Research for the Foundation Bettencourt Schueller
It’s a numbers game
To understand the connection between variation in the genes and variation in phenotype a vast amount of data must be gathered. For his 2018 Nature paper, Gianni gathered over 1000 isolates to build his collection (Peter & De Chiara et. al., 2018, Nature). These had to be sequenced, and then screened over 50 agar conditions before the phenotypes could be analysed. Gianni collaborated closely with Dr Joseph Schacherer to screen the collection in an experiment specifically designed for ROTOR+. Following the success of that paper (now over 1000 citations!) Gianni frequently replicates the library to distribute the collection to other labs world-wide.
For his next project, Gianni intends to understand the effect of inter-species interactions on the genotype/phenotype variation. The Liti lab has collaborated with 50 labs to bioprospect at 200 sites across Europe and North Africa. This led to the isolation of over 2000 novel yeast strains and other bacterial species from the oak microbiome. Gianni is now following the same game plan to characterise the interactions of over 3400 strains through his high-throughput automated platform.
“The ability to scale up from 10 to 100 to 1000 samples has changed the outcome and insights of our experiments completely”

Dr Gianni Liti | Leader of the Liti Lab
Institute for Research on Cancer and Aging, Nice
Isolate > Identify > Characterise > Repeat
Gianni stated that he continues to choose Singer Instruments for our “premium experimental tools” that he has seen consistent evidence for throughout the years. With ROTOR+, PhenoBooth+ and SporePlay+ now in his lab, Gianni is realising his automated experimental dreams.
“The ability to scale up from 10 to 100 to 1000 samples has changed the outcome and insights of our experiments completely”
Gianni describes ROTOR+ as a “game changer” when handling huge libraries of isolates. With ROTOR+, Gianni can replicate each strain across 100’s conditions to test for phenotypic differences. Gianni reminisced on his past work with Ed Louis, how installing a ROTOR was “really key to the development of their lab and their workflows”.
What is screened must be analysed. Gianni uses the PhenoBooth+ to image and analyse thousands of colonies each experimental round. PhenoBooth+’s ability to capture fluorescence is essential for Gianni’s study of this microbial community. Gianni described SporePlay+ as “really essential to [their] work, ultra-useful”. SporePlay+ is critical for studying the yeast life cycle, allowing Gianni to manipulate individual yeast spores or cells.

Automation is essential to expansion
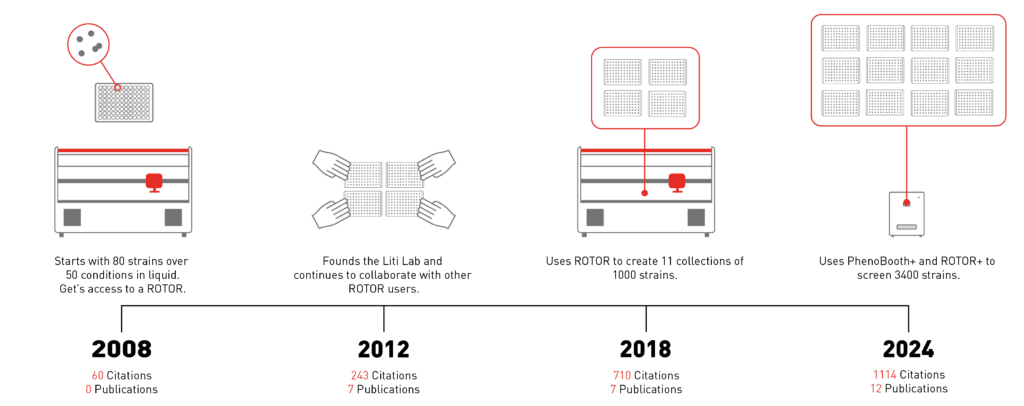
High-throughput genetic screening has fundamentally changed the way Gianni experiments. Researching the connection between variation in genotype and phenotype requires increasingly large strain libraries. Scaling up the numbers was critical for Gianni to understand this link. Gianni cited both ROTOR+ and Phenobooth+ as essential to making the transition from 100 to 1000 strains and now more.
“The ROTOR+ was key to the development of this workflow. The ability to manage this collection of over 3000 isolates is a gamechanger.”
Integrating Machine Learning
His genome-wide association studies use statistics to connect gene and phenotype together. Branching out into machine learning seems like the next logical step. Gianni’s lab recently trained a machine learning model on the data collected by an ensemble of study on the 1011 collection to predict phenotypes based on genetic sequence (Khaiwal et. al., 2025). The study provided important insights on how AI can provide novel information on the genotype-phenotype map. It also demonstrated how such approaches can be even more powerful if sample size is expanded.
Gianni now applies this model to his current collection of 3400 isolates where over 200 phenotypes are possible. Machine learning will allow Gianni and his team to integrate a more complex multi-omics approach to understanding variation by overlaying data on environmental conditions and microbiome interactions. Between Gianni’s automated workflow increasing his sample size, and his progress with machine learning, the Liti lab are drastically improving the efficiency of phenotype prediction.

Credit: Alexandre Darmon, Art in Research for the Foundation Bettencourt Schueller
Gianni reflects on automation
Gianni has worked synergistically with automation for a long time and has watched as it develops. “The scale that automation has reached is unbelievable. It has revolutionised the way we think and experiment”. Gianni said that just having a ROTOR+ in their lab inspired new ideas.
High-throughput automation allows Gianni to reach the current peak of understanding in genetic variation; he now believes that “it’s no longer a field of interest, but a genetic tool”. However using genetic variation as a tool to understand complex cellular processes is only possible with huge sample sizes. To manage an experiment harnessing the power of natural genetic variation, automation is essential.
Looking to automate large-scale genetic screens?
The ROTOR+ supports scientists to find that 1 colony in a million, in one day.
That’s why we have 10,000 citations and counting

Fiona Kemm | Marketing Coordinator
Fiona plays a key role in ensuring our innovations perform as intended, while providing exceptional support to users. Her efforts in the lab contribute to the reliability of our robots, developing protocols and assays that demonstrate their unique capabilities. She has a BSc in Biochemistry, an MRes in Molecular Microbiology and a talent for BioArt.