Serial dilution of live cells using SQWERTY
Aim
The aim of this experiment was to create a serial dilution of S. cerevisiae and E. coli using the pipetting robot SQWERTY and demonstrate the cell count is high enough for PIXL to pick from. The results were compared with a replicate using a Gilson manual pipette.
Method
Overnight cultures of S. cerevisiae and E. coli were grown in YPD and LB, respectively. A Gilson single-channel P200 & P20 and SQWERTY were used to perform the serial dilutions. Both pipettes were calibrated and 200 μl tips were used to complete the serial dilutions into a 96 multi-well plate (MWP). This was repeated in triplicate for each combination of organism and method.
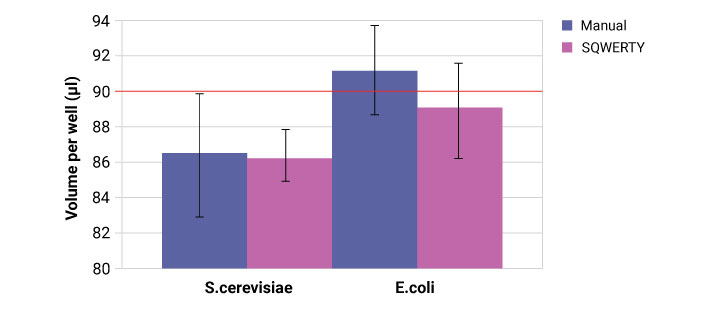
Weight change was calculated to estimate the average volume per well over the entire 96 MWP.
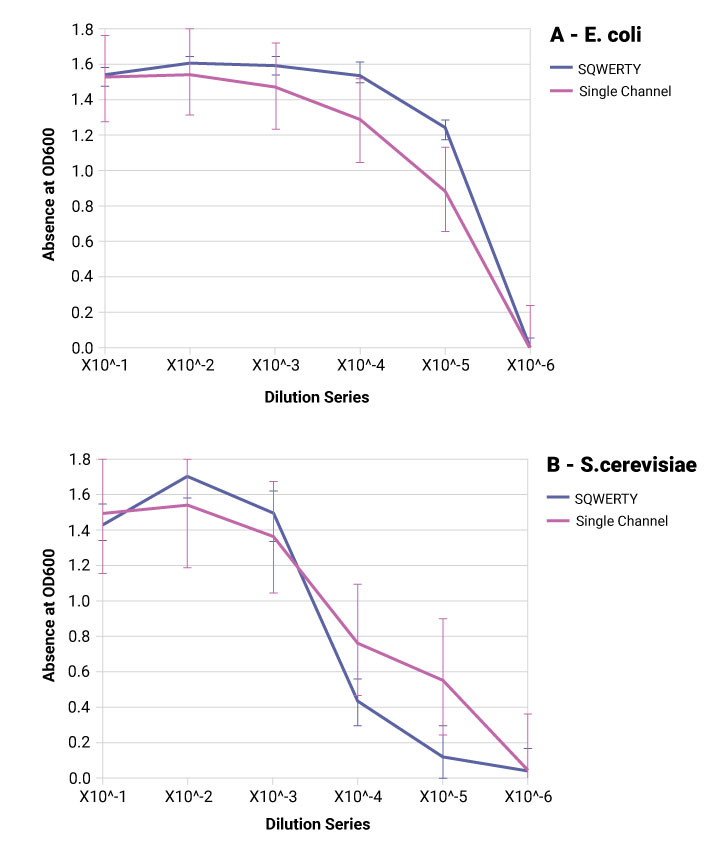
The absorbance at 600 nm (OD600) was measured with a SpectroStar Nano (BMG Labtech) spectrometer corrected against blank controls. This was performed after 24-hour incubation at 30 °C (120 rpm) to allow a detectable change. Average absorbance at each dilution step will show the success of serial dilutions and the standard deviation shows the consistency of volume transferred into each well (both calculated with MARS software).
Serial dilution using SQWERTY
A thorough clean was performed and a 20 min UV cycle was run to reduce the risk of external contamination during the experiment. The serial dilution operation was used to dilute a 2 mL Eppendorf tube of culture with 2x 2 mL Eppendorf tubes of broth in 4 replicate 1/10 serial dilutions to a final volume (at the moment of diluent dispense) of 100 μL. This was repeated for 3x 96 MWP (12 dilutions series). This was done using the built-in default water liquid class and advanced parameters (5 μL ‘Blowout’ and 1 μL ‘Travel Air Gap’ were enabled).
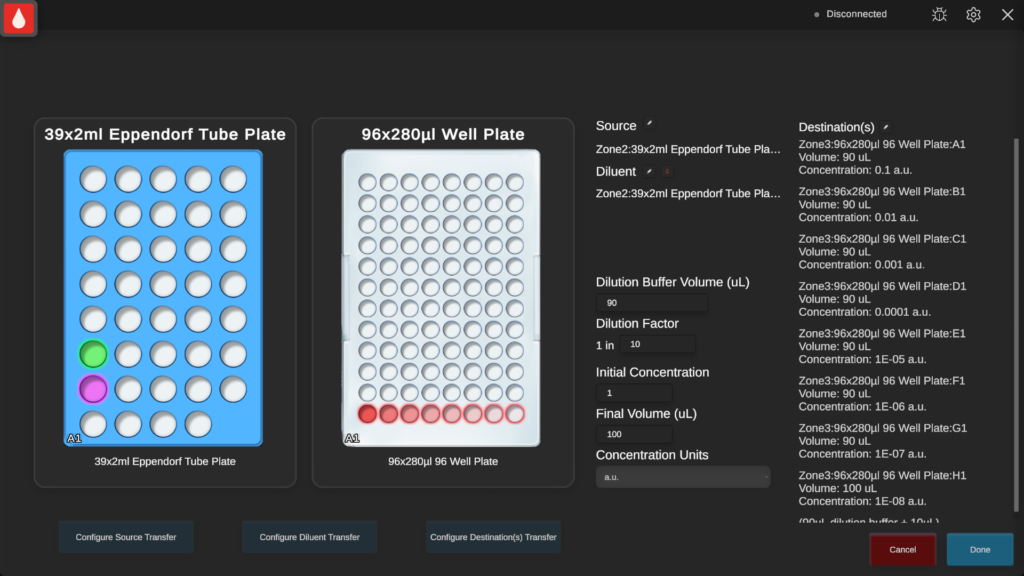
Serial dilution using a manual pipette
A P200 was used to transfer 90 μL sterile broth to each well in 4 columns, tip ejected every 8 wells to match the SQWERTY workflow. A P20 was used to transfer 10 μL culture between each well sequentially A-H. A mix was performed by circling & pipetting up and down 10 μL for each well. In SQWERTY, a mix with 90 μL was performed, but this can’t be replicated with the P20. The tip was ejected prior to each new well. This created a serial dilution across 8 wells by a factor of 1/10 and was repeated for 4 columns.
Results
Are the cells still alive?
It’s no use creating a serial dilution of cells if they are immediately sheared to death by less than gentle pipetting. Rumour has it this does happen in automated liquid handlers and pipetting robots.
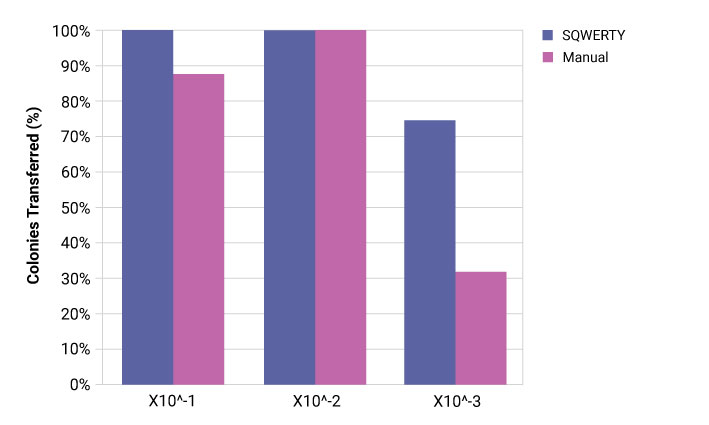
The PIXL re-array mode was used (default mix settings) to pick from each E. coli well 4 times to an LB agar plate immediately after serial dilution. This was then incubated overnight at 30 °C. The average number of colonies present was counted per dilution step to calculate the percentage of successful picks at each dilution step. Only E. coli was used as these cells are more likely to be damaged or killed by shearing during the pipetting process.
Summary
SQWERTY is able to produce a more consistent dilution series of both E. coli and S. cerevisiae compared to the manual single channel pipette. Each dilution series is more comparable to its replicates. The cells are alive enough to grow overnight be efficiently transferred using the PIXL colony picker.
The volume accuracy of each well is comparable in both SQWERTY and the manual pipette. The reason for the discrepancy between organisms is likely due to the fluid dynamics of yeast culture compared to E. coli culture and should be optimised using the advanced parameters and liquid classes on SQWERTY.
Trial & error and pipetting settings optimisation is essential for users of automated liquid handlers and pipetting robots. Fortunately, SQWERTY has the ‘Interactive’ mode for making quick adjustments to pipetting settings and witnessing the results in real-time with no workflow programming necessary. If you’re still struggling getting the most out of SQWERTY, our in-house scientists are available with advice through basic and advanced application support.

Fiona Kemm MRes | Scientist
Fiona is a vital member of our Research team, rigorously testing our robots to ensure scientists don’t break them. With no prior robotics experience, she was the ideal guinea pig for our world-class user experience and support. Holding a BSc in Biochemistry and an MRes in Molecular Microbiology, Fiona brings extensive hands-on expertise she applies across departments, supporting both users and internal teams. From writing insightful web articles to specialising in SQWERTY, Fiona ensures our innovations perform flawlessly, helping customers focus on the creative and interpretive aspects of science that can’t be automated.