Automatic zone of inhibition detection and analysis with PIXL
Abstract
Zone of inhibition assays are a cornerstone of antimicrobial research. This white paper introduces an automatic ZOI detection and analysis functionality which has been developed onto Singer Instruments’ PIXL colony picking workstation. It empowers the detection, screening and picking of colonies of antimicrobial-producing microorganisms.
PIXL’s ZOI mode (P-ZOI) is an intuitive and efficient tool that reliably measures a range of ZOI and colony sizes from different species on Petri dishes and SBS plates. P-ZOI measurements of zone size compare favourably to manual measurements. P-ZOI offers significant advantages over manual measurement with enhanced data traceability, export of comprehensive feature metrics and can be used in conjunction with PIXL’s precise and multipurpose automatic colony picking capability. With applications in antimicrobial compound discovery, synthetic biology, and microbial ecology, P-ZOI offers users a reliable and versatile solution to streamline ZOI workflows, improve data acquisition, and accelerate downstream processes.
Overview
P-ZOI has an intuitive user experience which guides users through performing a zone of inhibition workflow. The workflow consists of imaging a 90 mm Petri dish or SBS plate in PIXL (Figure 1). PIXL then uses state-of-the-art colony and ZOI detection algorithms to segment the image to distinguish between the lawn, ZOIs and colonies. Users select which colonies and zones they wish to take forward for analysis or colony picking using filters common to other PIXL workflows. These filters can select for zones and colonies based on parameters including size, colour and proximity to other features on the plate. Once the user is happy with their selection, the quantified metrics of the ZOIs and colonies can be exported along with raw and annotated images of their plates. Users can then use PIXL to pick colonies of interest to different plates for further culturing in agar or liquid media or for downstream applications such as PCR.

Use PIXL to automatically detect colonies and ZOI on 90mm Petri dishes or SBS plates

Screen features are based on parameters such as size, colour and proximity to other features.

Export publication-quality annotated images and feature metrics.
[Optional] Use PIXL to pick colonies within zones for further culturing and other downstream applications.
Figure 1: Overview of performing a zone of inhibition workflow in PIXL.
Detect zones and colonies with ease with PIXL
The flexibility of P-ZOI empowers users to adapt the software to their specific requirements. Lighting conditions, detection algorithms, and filtering parameters can be adjusted, allowing for optimal zone and colony detection. P-ZOI incorporates advanced algorithms capable of detecting features with differences in colony morphology, lawn colour and texture, ZOI size and overlapping ZOIs. This adaptability ensures reliable performance when working with diverse microbial species or complex experimental conditions (Figure 2).
P-ZOI has been validated to detect central colonies within zones with diameters of 2 to 9 mm, and can detect zones between 8 and 80 mm with up 40% overlap (by radius) with other zones.
By automating the detection of zones of inhibition, PIXL eliminates the need for manual measurements, minimising the potential for human error and allowing for better traceability between samples and analysed data.
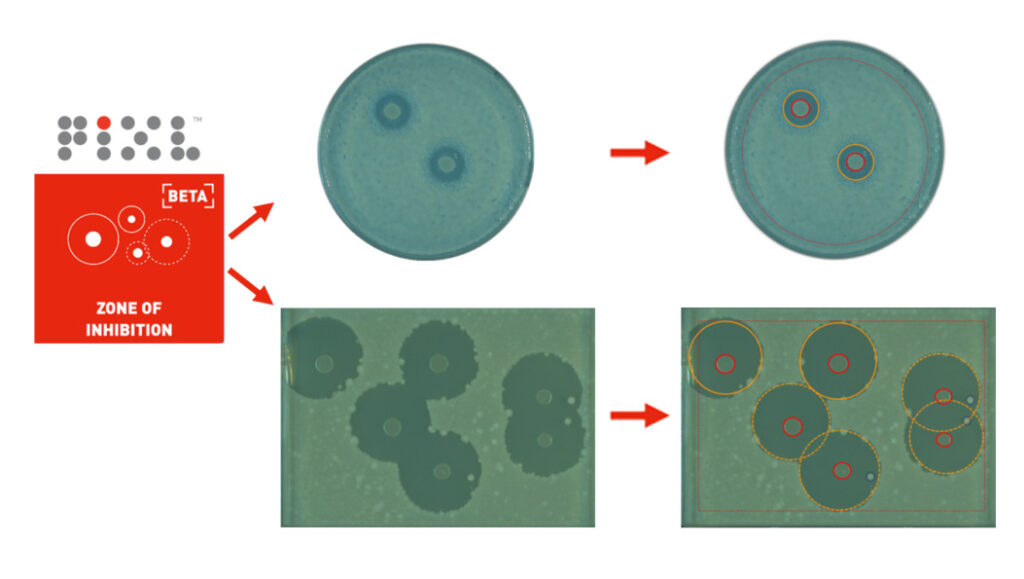
Figure 2: P-ZOI can be applied to a range of experimental requirements. (Above) Automatic zone and colony determination on 90 mm petri dishes with Escherichia coli lawn and central colonies. Zones of inhibition are circled in yellow. Colonies are circled in red. (Above) Automatic overlapping zone and colony determination on SBS plates with Bacillus subtilis lawn and E. coli central colonies. Overlapping zones are highlighted with yellow dashed lines. Central colonies have been screened using PIXL’s centrality filter; the smaller, non-central colonies are detected and annotated with a blue outline.
PIXL exports measurements and images for downstream applications
The output of P-ZOI provides users with comprehensive tables and annotated images for analysis and publication purposes (Figure 3). The exported tables contain quantitative measurements of feature parameters such as colony and ZOI size and proximity to other features and semi-quantitative measurements such as colony brightness. These metrics enable users to perform statistical analysis, compare results across experiments, and gain valuable insights from their data.
Accompanying the tables, PIXL generates annotated images that can be cross-referenced against the data tables. The annotated image can be exported with colony ID, ZOI ID or ZOI radius displayed or hidden as required. These images serve as visual references, facilitating result sharing and verification, and enables data traceability. Un-annotated images are also exported which allows users to perform their own analysis or annotation in third-party software.
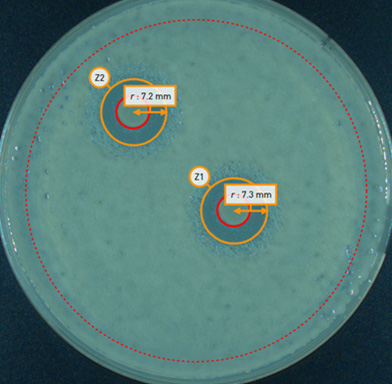
| Colony Name | Diameter | Distance from zone centre (mm) |
| Z2C1 | 7.4 | 0.3 |
| Z1C1 | 7.3 | 0.2 |
| Zone Name | Diameter | Zone overlap (%) |
| Z2 | 14.658 | 0 |
| Z1 | 14.658 | 0 |
Figure 3: P-ZOI exports data for downstream applications. (left) Annotated ZOI plate automatically generated by PIXL. Labels in the image correspond to exported (right) tables which contain measurements of detected features.
Pick colonies within zones of inhibition using PIXL
PIXL offers a practical and efficient solution for researchers to pick colonies growing within zones of inhibition and transfer them to different plates for further experimentation.
With PIXL, users have the flexibility to pick colonies within zones of inhibition and transfer them to various plate formats, including fresh agar plates, multiwell or deep well plates containing media or assay reagents, or PCR plates. This feature expands the range of possibilities for subsequent culturing, screening, and characterisation of selected colonies.
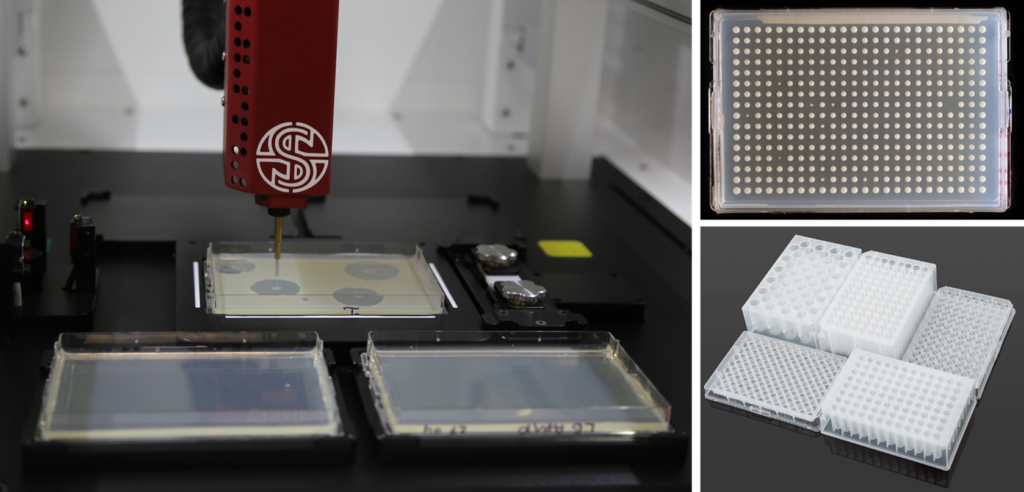
Figure 4: PIXL can precisely pick colonies within ZOIs and transfer material to fresh agar plates or a variety of SBS-style plates for further culturing or for downstream assays such as PCR.
PIXL performs ZOI workflows reliably and streamlines downstream applications
Manual measurement of ZOI radius can be subjective due to the irregular shape of zones and the presence of growth gradients between the lawn and zone boundary. In our observations, we found significant variation in manual measurements of ZOI radius among different users, with a standard deviation (SD) of 0.5 mm. Over 80% of PIXL’s radius measurements were within 2 SD of the manually determined mean radius, demonstrating a high degree of similarity between PIXL’s output and manual measurements.
One significant benefit of using PIXL for ZOI workflows is the storage and traceability of data. PIXL stores all generated data, allowing researchers to easily trace back measurements to specific zones or colonies on specific plates. This feature enhances data management and facilitates result verification and reproducibility. Additionally, PIXL’s data export functionality includes a wide range of metrics that are not easily measured with manual methods, such as colony and zone colour, brightness, circularity, and overlap with other zones. This comprehensive data provides users with more opportunities to generate insights into interactions between microorganisms on their plates.
A key advantage of performing ZOI workflows with PIXL is that downstream processing of colonies with zones is made easier with automatic colony picking. PIXL accurately determines the positions of colonies within zones, with a positional accuracy of 0.13 mm (± 0.05 SD) using default conditions. This precision enables PIXL to precisely pick colonies and transfer them to new plates for downstream applications. By automating the colony-picking process, PIXL saves researchers time and effort and removes the opportunity for human error.

Applications
PIXL ZOI mode has the potential to be used in a broad range of applications.
Antimicrobial compound discovery workflows
PIXL’s ZOI mode can be utilised to screen microbial libraries or environmental samples and isolate colonies that produce antimicrobial compounds against target microorganisms.
For example, environmental samples taken from soil could be screened on the lawns of a target microorganism. After incubation, PIXL’s ZOI mode could be employed to detect and measure the zones of inhibition surrounding colonies which are producing antimicrobials against the target microorganism.
These colonies can then be prioritised based on measured properties of colonies and zones of inhibition (e.g. ZOI size). PIXL can then be employed to pick colonies of interest for downstream processing to accelerate the characterisation of microbial strains with antimicrobial activity against the target of interest.
Synthetic biology and metabolic engineering workflows
PIXL’s ZOI mode can be utilised to screen libraries of genetically modified strains or mutant libraries based on measured properties of colonies and zones of inhibition.
For example, PIXL’s ZOI mode can be used to detect zones of inhibition surrounding colonies engineered to produce a particular compound of interest. By quantifying the size and shape of the zones, users can identify colonies with the highest antimicrobial activity.
These colonies can then be picked with PIXL for further characterisation and optimisation to accelerate the development of microbial strains with improved production capabilities.
Microbial ecology workflows
PIXL’s ZOI mode can be utilised to assess the interactions within microbial communities, helping unravel complex ecological relationships.
For example, PIXL’s ZOI mode can be used to study the interactions between pathogenic and commensal bacteria isolated from the human gut. By analysing the zones of inhibition on agar plates, users can determine the inhibitory effects of pathogenic strains on commensal microbes or vice versa.
Implementing your own ZOI workflows
Book a 30-minute slot to discuss your workflow with one of our product experts,
or ask us anything. 1700+ happy microbiologists and counting.