A protocol for Chlamydomonas mating, sporulation, dissection and analysis.
Industrial solutions from academic research
Fiona Kemm1 and Dr Michal Breker-Dekel2
1 Singer Instruments, 2 The Hebrew University of Jerusalem
Chlamydomonas Genetic Research
Dr Michal Breker-Dekel, assistant professor at the Hebrew University of Jerusalem, is an early pioneer of genetic research in Chlamydomonas reinhardtii, having established herself mapping genetic interactions in a plant species. Building on equivalent approaches in Saccharomyces cerevisiae, her work has played a major part in mapping essential genes, helping to establish this unicellular green algae as a powerful microbial model organism for multicellular land plant cell biology (Breker et. al., Plant cell 2018).
A fundamental understanding of the genes involved lets the Breker lab interrogate the systems biology of chloroplast metabolism, with the goal of producing bio-resilient plants that can survive our changing climate.
Tracking genetic inheritance through tetrads
Tetrad dissection serves as an invaluable tool for validating and refining hits identified in large-scale genetic screens. Analysing the ratios of meiotic products provides Michal with vital information about the nature and mechanism of any genetic interactions detected. Without specialist solutions this is time-consuming and error prone to do at scale, especially considering the delicate nature of Chlamydomonas zygospores.
SporePlay+, Singer Instruments’ best-selling tetrad dissection workstation, was originally developed for dissecting the robust and relatively indestructible spores of yeast. But in 2021 Michal’s lab began experimenting with its use in C. reinhardtii. In this application note, Michal shares her positive results, detailing the methods and top tips her lab has developed to achieve success.
Key advantages of SporePlay+ for Chlamydomonas tetrad dissection
- Dissect up to 200 tetrads per day, arrayed in grids of up to 20 per plate.
- Quick, precise micromanipulation, ensuring maximum survivability of delicate progeny.
- Precise clickstops to help easily position and track cells of interest.
Tetrad Dissection in Chlamydomonas research
To better understand gene function, Michal creates libraries of up to 200,000 Chlamydomonas cells and then systematically analyses them in two independent screens to identify which colonies contain mutations that prevent a function within the chloroplast (e.g. protein quality control).
The next step is to use SporePlay+ to establish whether the mutant phenotype is caused by one driving mutation or a combination. SporePlay+ is an all-in-one solution to Chlamydomonas tetrad dissection, a simple but precise technique for tracking genetic inheritance.
The ability to directly track the inheritance of alleles from the mating pair through to the progeny is essential to draw strong conclusions about genetic linkage in Chlamydomonas. “Without tetrad dissection you couldn’t be certain. When a detailed analysis of quantitative allele segregation patterns is required, SporePlay+ excels,” says Michal.

Figure 1: SporePlay is robust, works well and is easy to use. Photo credit – Noemie Alon
Protocol
Chlamydomonas mating cross
- Suspend a fresh mutant query colony in nitrogen free gamete induction medium (Dutcher, Methods Cell Biol. 1995). Repeat with a culture of the opposite mating type in a different vessel. The OD 750 should be ~0.2.
- The mating types are mt+ or mt-.
- Incubate both vessels for 5 hours under LED bulbs (1000 lux) at 21°C to induce gametogenesis (Breker et. al., JOVE 2016).
- Mix the mutant and WT suspensions together and allow to mate for 30 minutes in liquid under light.
- Plate the mixed culture onto a Tris-Acetate-Phosphate (TAP) agar PlusPlate. Incubate at 21 °C under light overnight. Then wrap in foil to continue incubating in the dark for a further 5 days. This allows the diploid zygospore progeny to form.
- Use a scalpel to clean up any vegetatively growing cells. Scratch the agar lightly and pick the zygospores using a cut tip and 100 μl TAP.
- Streak a line of the zygospores onto the edge of a TAP agar 90 mm petri dish.
- Return the plates to the light to allow meiosis and formation of the tetrad over the next few days.
Chlamydomonas tetrad dissection
- Place the petri dish, agar facing down, onto the SporePlay+ and maneuver the stage to see the sample streak using the eye pieces (or the trinocular head camera).
- Unlike yeast, the C. reinhardtii tetrad ascus breaks automatically so not all tetrads will be ready for dissection at the same time.
- Pro tip from Michal: Keep returning the plates to SporePlay+ every hour or so, to locate and dissect any collapsed tetrads while you do other project work.
- Use the needle to gently separate the haploid cells from each other and place each cell at sequential click stops in either rows or columns. Glance at the index pointer to help you return to the correct positions as you move between the sample and the matrix.
- Pro tip from Michal: Chlamy cells don’t appreciate agitation and
won’t survive off of the agar for more than 20 seconds. So make sure
that you move each cell quickly but gently.
- Pro tip from Michal: Chlamy cells don’t appreciate agitation and
- Once all the tetrads are dissected, return the plate to incubate in the light at 21°C for 7 days.
Analysis & downstream processing
- Test the dissected tetrads for a mutant or WT phenotype using your preferred assay, e.g. presence of fluorescence or difference in colony size.
- If two of the segregates show the mutant phenotype and two show the WT phenotype then the mutant phenotype is driven by a mutation in a single gene (see Figure 2).
- Having identified the mutants of that interest, a PIXL colony picker is then used to put them into re-array format for further processing or long term storage.
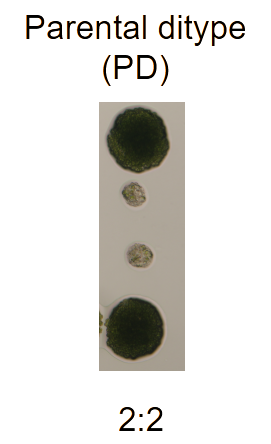
Figure 2: The parental ditype segregation pattern of a mutant x wild type
C. reinhardtii mating cross.

“Chlamydomonas cells are very sensitive and won’t survive long off of the agar. But most of my students can master the tetrad dissection technique within a couple of mating crosses. SporePlay+ is stable, robust, easy and works well.”
Dr Michal Breker
The Breker Lab, Hebrew University of Jerusalem
Performing precise tetrad dissection?
Reliable and trusted by labs worldwide – easy to use and work with the most delicate organisms.

Fiona Kemm | Research Scientist
Fiona plays a key role in ensuring our innovations perform as intended, while providing exceptional support to users. Her efforts in the lab contribute to the reliability of our robots, developing protocols and assays that demonstrate their unique capabilities. She has a BSc in Biochemistry, an MRes in Molecular Microbiology and a talent for BioArt.