Age Analysis
Jan Singer. Singer Instruments, Roadwater, Somerset, UK, TA23 0RE
Replicative Lifespan Analysis
Replicative lifespan (RLS) analysis is a vital tool for yeast geneticists interested in ageing studies. The replicative lifespan of a yeast cell refers to the number of daughter cells one mother cell can produce before becoming senescent. To do this a virgin mother cell is identified and selected. The number of daughter cells produced is then counted until the mother cell becomes exhausted.
Historically, performing RLS analysis was cumbersome and difficult because it involved a high level of manual dexterity and skill. In recent years the technique has become easier, largely due to the development of easy-to-use micromanipulation systems, programmed aging protocols and the ability to record still and time-lapse images.
Principles
A mother cell is selected from an inoculum on one side of an agar plate and isolated. It is allowed to bud and carefully watched until separation occurs. (Figure 1a) The mother cell is then carefully removed leaving behind a daughter cell which is a virgin mother. The daughters from the virgin mother are carefully removed (Figure 1b). The process is repeated until the virgin mother becomes senescent.
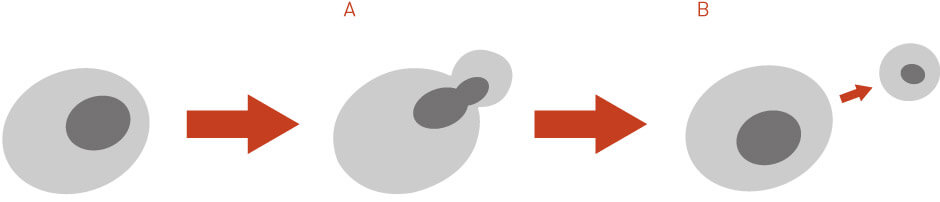
Figure 1 A: Diagram illustrating Yeast Cell Budding. See text for details.
Figure 1 B: Diagram illustrating Daughter Cell Discard. See text for details.
Mutations, chemical stressors and drugs may increase or decrease the replicative lifespan of yeast cells. This can be useful, for example, in determining the biological processes that govern ageing and ageing-related diseases in humans.
Theory
Yeast is a great model for understanding the process of ageing because its lifespan can be measured through the RLS analysis. RLS analysis takes advantage of the fact that yeast cells divide by asymmetric budding and produce a daughter cell that is much smaller than the mother. Researchers can count the number of daughter cells as they separate the daughters away from the mother, thereby determining the lifespan of the mother cell.
Selection
Cells are plated onto an agar dish or agar-coated glass slide. This is inverted onto a specialized microscope workstation that is designed for mother-daughter separation or ageing analysis. These microscopes incorporate a micromanipulator device so the user can pick cells from the agar surface, place the cells in a predetermined orthogonal array and separate the daughter cells as desired, counting the number of divisions and discards.
Application Age Analysis Extrachromosomal rDNA Circles— A Cause of Aging in Yeast
Abstract (David A Sinclair, Leonard Guarente)
Although many cellular and organismal changes have been described in ageing individuals, a precise, molecular cause of ageing has yet to be found. A prior study of ageing yeast mother cells showed a progressive enlargement and fragmentation of the nucleolus. Here we show that these nucleolar changes are likely due to the accumulation of extrachromosomal rDNA circles (ERCs) in old cells and that, in fact, ERCs cause ageing. Mutants for sgs1, the yeast homolog of Werner’s syndrome gene, accumulate ERCs more rapidly, leading to premature ageing and a shorter life span. We speculate on the generality of this molecular cause of ageing in higher species, including mammals.
Regulation of Yeast Replicative Life Span by TOR and Sch9 in Response to Nutrients
Abstract (Matt Kaeberlein, R. Wilson Powers III, Kristan K. Steffen, Eric A. Westman, Di Hu, Nick Dang, Emily O.Kerr, Kathryn T. Kirkland, Stanley Fields, Brian K. Kennedy)
Calorie restriction increases life span in many organisms, including the budding yeast Saccharomyces cerevisiae. From a large-scale analysis of 564 single-gene–deletion strains of yeast, we identified 10 gene deletions that increase replicative life span. Six of these correspond to genes encoding components of the nutrient-responsive TOR and Sch9 pathways. Calorie restriction of tor1D or sch9D cells failed to further increase life span and, like calorie restriction, deletion of either SCH9 or TOR1 increased life span independent of the Sir2 histone deacetylase. We propose that the TOR and Sch9 kinases define a primary conduit through which excess nutrient intake limits longevity in yeast.alorie restriction increases life span in many organisms, including the budding yeast Saccharomyces cerevisiae. From a large-scale analysis of 564 single-gene–deletion strains of yeast, we identified 10 gene deletions that increase replicative life span. Six of these correspond to genes encoding components of the nutrient-responsive TOR and Sch9 pathways. Calorie restriction of tor1D or sch9D cells failed to further increase life span and, like calorie restriction, deletion of either SCH9 or TOR1 increased life span independent of the Sir2 histone deacetylase. We propose that the TOR and Sch9 kinases define a primary conduit through which excess nutrient intake limits longevity in yeast.
Genetic analysis of ageing: role of oxidative damage and environmental stresses
Abstract (George M. Martin, Steven N. Austad & Thomas E. Johnson)
Evolutionary theory predicts substantial interspecific and intraspecific differences in the proximal mechanisms of ageing. Our goal here is to seek evidence for common (‘public’) mechanisms among diverse organisms amenable to genetic analysis. Oxidative damage is a candidate for such a public mechanism of ageing. Long-lived strains are relatively resistant to different environmental stresses. The extent to which these stresses produce oxidative damage remains to be established.
An Age-Induced Switch to a Hyper-Recombinational State
Abstract (Michael A. McMurray, Daniel E. Gottschling)
There is a strong correlation between age and cancer, but the mechanism by which this phenomenon occurs is unclear. We chose Saccharomyces cerevisiae to examine one of the hallmarks of cancer—genomic instability— as a function of cellular age. As diploid yeast mother cells aged, an ∼100-fold increase in loss of heterozygosity (LOH) occurred. Extending life-span altered neither the onset nor the frequency of age-induced LOH; the switch to hyper-LOH appears to be on its own clock. In young cells, LOH occurs by reciprocal recombination, whereas LOH in old cells was nonreciprocal, occurring predominantly in the old mother’s progeny. Thus, nuclear genomes may be inherently unstable with age.
MSM 400 is undeniably the most powerful tetrad dissection workstation in the World. The built-in software contains useful protocols that automate many of the repetitive aspects in dissection, ageing, and screening studies.
Find out more about the MSM 400
See how you can improve your scientific research with the most powerful
tetrad dissection workstation in the World.