High-throughput Cell Plating with ROTOR+
Jay Yang¹, Michaela Spitzer¹, Addison Wong², Wen Shan Yew³, and Matthew Chang³
¹Singer Instruments, Roadwater, TA23 0RE, UK
²Singapore Institute of Technology, 10 Dover Drive, Singapore 138682, Singapore
³Synthetic Biology for Clinical and Technological Innovation (SynCTI), Life Sciences Institute, National University of Singapore, Singapore 117456, Singapore
Introduction
Advances in robotic automation help standardise repetitive procedures in labs. This in return increases throughput and reliability. The plating and isolation of single colonies is indispensable in the course of any micro- and molecular biology study. Despite the clear benefits of automation, plating cells on robotic platforms is not widely adopted. This is largely due to the inconsistency and the low throughput of current robotic solutions – plating one sample at any one time. This is not practical when working with hundreds of thousands of samples at a time.
At Singer Instruments, we have developed a solution for this problem: the high-throughput plating protocol using the ROTOR+. This protocol provides researchers the capacity to simultaneously plate 96 samples in under two minutes on a single plate, a truly high-throughput approach saving valuable time and resources.
ROTOR+ Plates Cells Using a Custom Pinning Matrix
To take advantage of the ROTOR+’s liquid-to-agar transfer functionality, we have developed a plating protocol that uses 96-density long pin RePads to transfer cells from a 96-well microplate to an agar plate. For this protocol the ROTOR+ pins a 7×7 matrix for each of the 96 samples on a rectangular agar plate. Once the ROTOR+ has picked up samples from the 96-well plate, it repeatedly spots the pintip of the RePad onto the agar plate without revisiting the source plate. The repeated spotting of the pintips dilutes the cells on the pintips as the ROTOR+ progresses through the matrix, resulting in pinning of single colony-forming units (Figure 1).
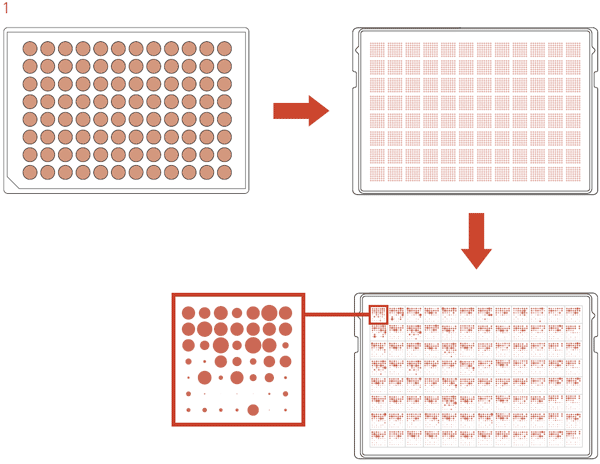
Figure 1: Schematic diagram illustrating the 7×7 matrix protocol.
The ROTOR+ pins samples from a 96-well source plate in the 7×7 matrix onto an agar plate. After incubation single colonies can be observed for all 96 samples.
The ROTOR+ Plating Protocol is Capable of Plating Yeast and Bacterial Cells
To assess the effectiveness of this protocol, we grew an overnight culture of S. cerevisiae, performed a 2-fold dilution series of the overnight culture, and pinned the cells onto PlusPlates (Figure 2a). After 48 hours incubation at 30°C we observed single colonies across a range of dilutions.
We repeated the same experiment with P. pastoris (Figure 2b) and E. coli (Figure 2c). The results were consistent and we observed growth of single colonies at various dilutions. This demonstrates that the 7×7 matrix plating protocol is applicable to a range of microorganisms, including yeast and bacterial cells.

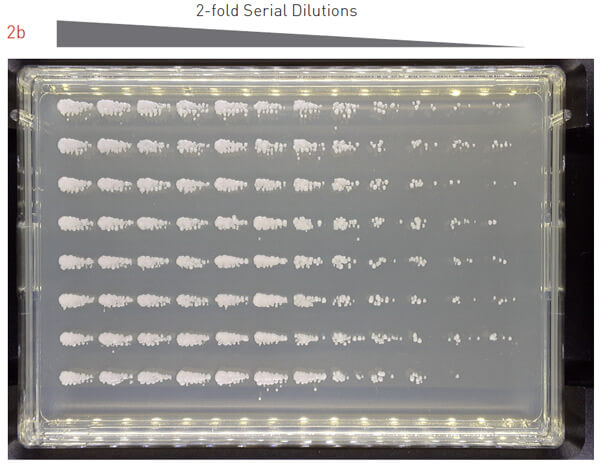

Figure 2: Testing the 7×7 matrix plating protocol with different model organisms.
Overnight cultures for different model organisms were 2-fold dilution series and plated onto an agar.
A) Plated S. cerevisiae was incubated at 30°C for 48 hours.
B) Plated P. pastoris was incubated at 30°C for 36 hours.
C) Plated E. coli was incubated at 37°C for 18 hours.
The ROTOR+ Prepares a Plate of 96 Samples of Yeast Cells
The 2-fold dilution series suggests that 1:16 to 1:128 dilutions result in single colonies for many samples. Therefore we repeated the experiment using a 1:32 dilution of a yeast overnight culture to confirm our conclusion. The resulting image (Figure 3) shows that every sample was clearly defined in its matrix with single colonies and without cross-contamination.
The ROTOR+ is the fastest and most powerful colony manipulation robot in the world. It is essential for high-throughput screening and single colony picking.
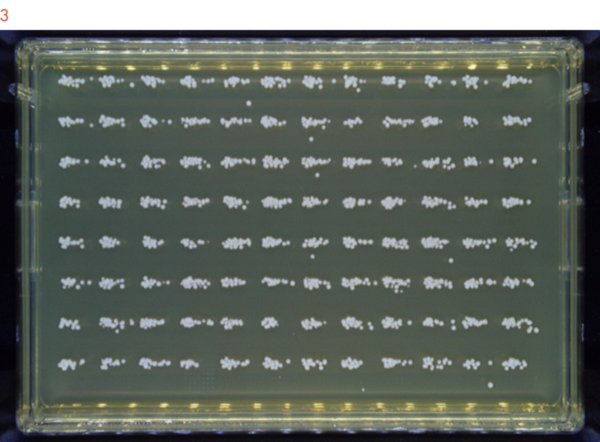
Figure 3: Samples are plated without cross-contamination.
A S. cerevisiae overnight culture was diluted 1:32 and plated onto agar. The plate was incubated at 30C° for 36 hours.
The Protocol can Plate Cells From E. Coli Transformation
E. coli transformation is the most common protocol where large numbers of transformants need to be plated to pick single colonies. To determine whether the ROTOR+’s 7×7 pinning protocol is able to plate E. coli cells after transformation, we transformed E. coli with either a) a plasmid encoding GFP, b) an empty vector, or c) dH2O without any plasmid. Our data indicate that we were able to obtain single colony-forming units on LB + Kan plates when cells were transformed with plasmids, while the cells transformed without plasmids were not viable and did not grow on the selective media (Figure 4a).
To test the specificity of the transformation, we imaged the plate using the PhenoBooth, which is able to detect GFP signals. As expected, the cells that were transformed with GFP plasmid showed strong GFP signals, while the cells that were transformed with empty vector did not (Figure 4b).
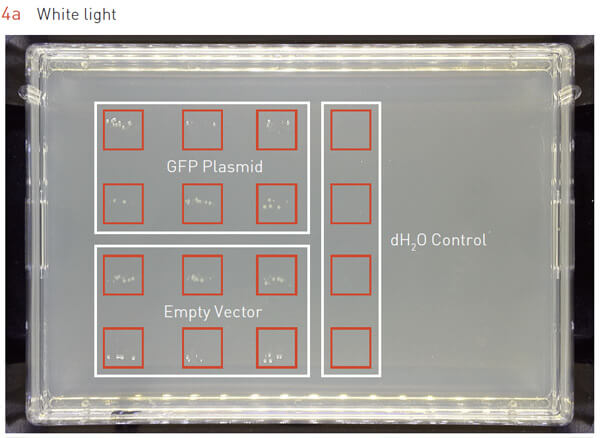

Figure 4: 7×7 matrix protocol is able to plate E. coli transformants.
E. coli cells were transformed with either GFP-vector, empty vector, or water. After transformation, the 7×7 matrix protocol was used to plate these cells for single colonies.
A) Cells transformed with either GFP-vector or empty vector formed colonies on the plate, while cells transformed with water failed to form colonies.
B) The plate was imaged with the PhenoBooth using UV light to detect GFP signal. Cells transformed with GFP-vector formed colonies with a GFP signal, while cells transformed with empty vector failed to fluoresce.
Conclusion
The ROTOR+ 7×7 matrix protocol automates high throughput plating of microbial organisms for single colonies. This protocol allows researchers to streak 96 samples onto one target plate in less than two minutes, using a single 96-long pin RePad. Altogether, the ROTOR+ provides a truly high-throughput and low-cost solution that merely takes 1.25 seconds and costs 4.2 cents per sample. Our cutting-edge protocol will save time, resources, and money.
This new matrix plugin for the ROTOR+ provides a protocol that adds significant value and flexibility to an already robust system. The default 7×7 matrix is fully customisable as is the location where the matrix is spotted. If you are working with a different model organism and would like to be the first to test this pioneering technology, please contact us.
Streamline your workflows with user-friendly lab automation
ROTOR+ lets you simultaneously streak 96 samples on a single plate in under two minutes.