Lactobacillus in probiotics and food production
What is Lactobacillus and how is it used?
Introduction
Lactobacillus bacteria are naturally found in the human body as part of the gut microbiota, as well as in fermented foods such as yogurt, cheese, and sauerkraut. The genus includes L. bulgaricus, L. rhamnosus, L. sakei and at least 258 other species (Zheng et al, 2020).
Common to all Lactobacillus bacteria is the production of antimicrobial compounds, such as lactic acid, hydrogen peroxide, bacteriocins, and organic acids, as a metabolic byproduct of fermentation. This has led to Lactobacillus bacteria being widely studied for their health benefits, both as a probiotic and as a food preservative to reduce risk of food borne illness.
Animal feed

Lactobacillus-based probiotics are used to promote gut health and improve the performance of livestock animals, leading to better growth rates, feed conversion efficiency, and overall health of the animals.
Food preservation

Lactobacillus strains can help suppress the growth of spoilage microorganisms and harmful bacteria, allowing foods to be stored for longer without the need for refrigeration or chemical preservatives.
Immune support

Lactobacillus helps maintain a balanced gut microbiota by regulating immune response and preventing overgrowth of harmful pathogens to reduce the risk of infections, allergies, and autoimmune diseases.
Strain development
Production strains of Lactobacillus are typically developed by means of an iterative process consisting of isolating and culturing wild type strains, reformatting isolates into microplates or microarrays for efficient sample handling, screening to assess sample fitness in various industrial conditions, analysis of results to select best candidates and finally directing positive hits into downstream pathways for scaling and production or, if there are none, then repeating the process (fig. 1).
The ability to efficiently create high density arrays from colonies growing randomly on agar can greatly streamline this process by enabling isolates to be efficiently re-arrayed and replicated to facilitate higher-throughput analysis (Aguiar-Cervera et al, 2021).
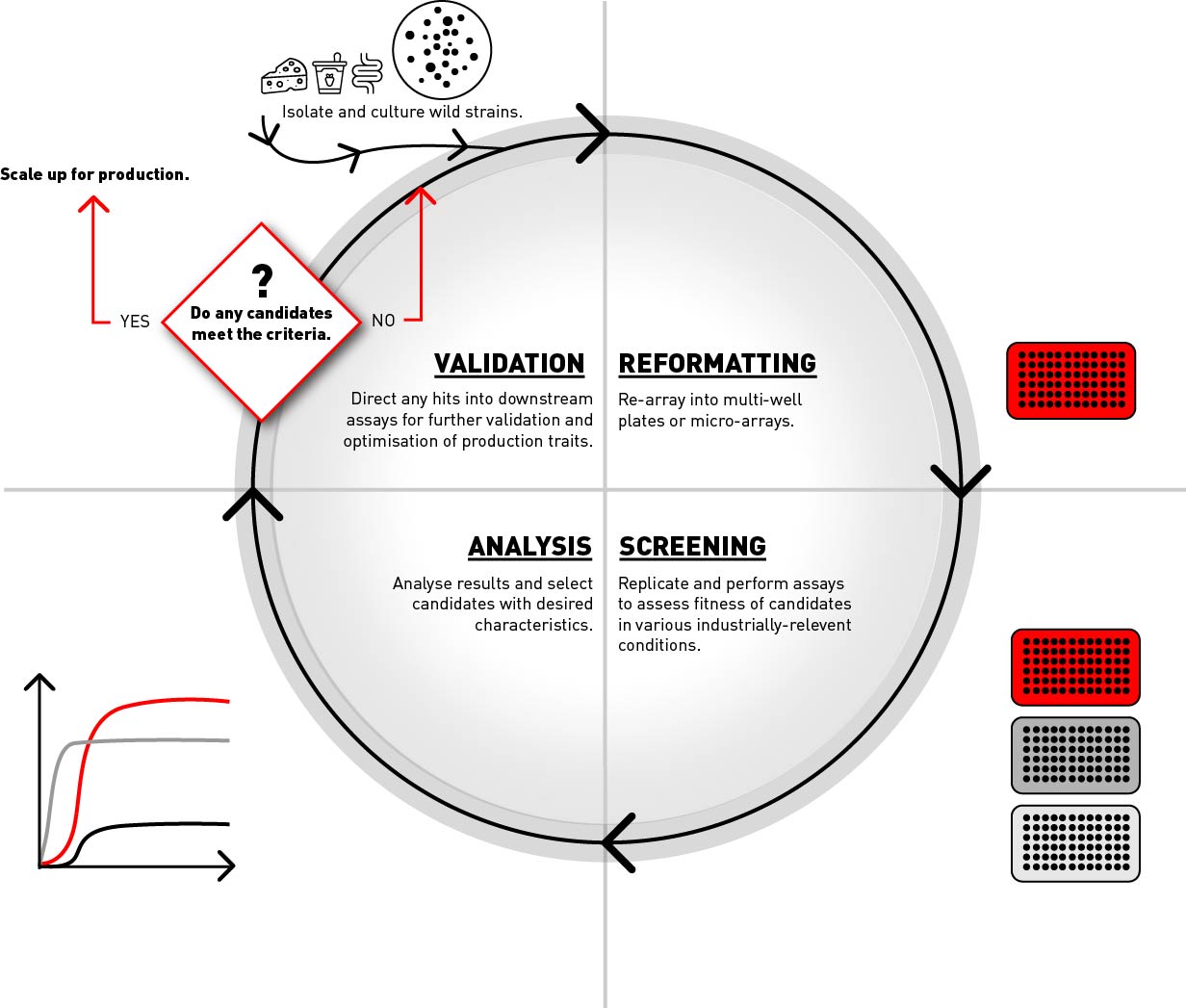
Figure 1: A high throughput screening workflow showing how Lactobacillus isolates can be re-arrayed and replicated in solid media to assess fitness within various industrial conditions.
The throughput challenge
Due to the vast variety of different species of Lactobacillus, identifying suitable strains and optimising them for specific industrial processes, such as animal feed or human probiotics, can be a long and complex process. The probability of finding a hit is proportional to its frequency in the population and the available screening capacity, with large libraries often needed to achieve results within commercially-competitive timelines.
Achieving sufficient throughput is further compounded by the following issues, particularly within labs reliant on manual sample handling and tracking:
Sample handling

Contamination from cleaning chemicals or mixing of colonies can be a significant source of error in labs that rely on manual colony picking. Ensuring each colony is picked and transferred to the appropriate growth medium or assay can be time-consuming and error prone.
Cost-effectiveness
Even skilled technicians need to work at lower densities to achieve consistency and accuracy when arraying microbial plates manually. This leads to increased demand for staff, labware, reagents and space, thereby inflating consumables budgets and overhead costs.
Data management

Unlocking the full potential of big data means screening vast libraries of strains. Tracking information efficiently across all those data points, such that they can be traced back to individual strains can be a huge challenge.
Robotic colony pickers
Robotic colony picking systems can drastically speed up the process of identifying promising probiotic candidates by enabling thousands of colonies to be screened accurately, reproducibly and at low cost.
Strains with desirable characteristics, such as high survivability in the gut, ability to produce beneficial metabolites, or antimicrobial properties, can be efficiently screened by programming robotic pickers to pick colonies based on specific criteria, including growth rate, response to different culture conditions, or measuring zones of inhibition (fig. 2).
In recent years, the development of ultra-high-precision colony pickers based on agar surface detection and contact pressure regulation has further increased the reliability of these systems, facilitating the analysis of vastly more strains in less time and with next to zero errors.

Figure 2: Some colony picking robots can help accurately measure zones of inhibition as part of screening assays to identify strains with the strongest antimicrobial properties.
“We are screening for strains that could be useful either as production strains or as probiotics.
The PIXL, we have so far used to spot our strains on 96 well agar plates for assaying.”

Dr Sandra Kittelmann,
Group Leader at Wilmar International.
Robotic colony pickers can facilitate development of production strains by enabling the quick, efficient screening of large numbers of candidates to identify those with desired characteristics, such as improved digestion, enhanced nutrient absorption, or immune modulation.
For example, Wilmar International are using PIXL to identify strains of lactobacilli picked from MRS agar to be cultured in bioreactors and supplied as probiotics to chickens (fig. 3).
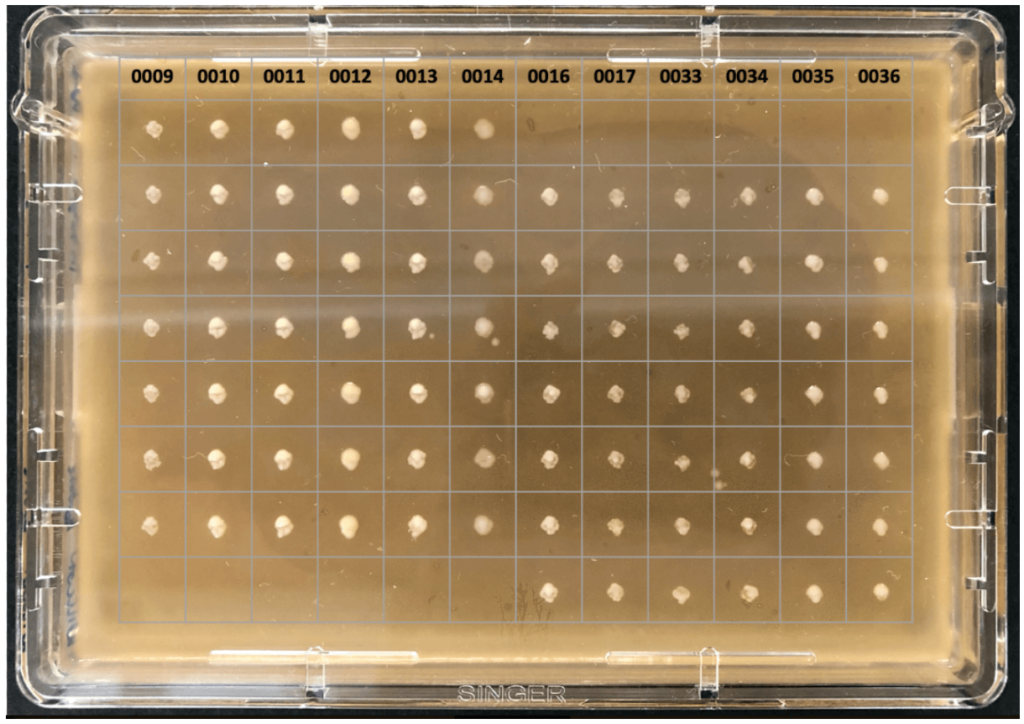
Figure 3: Lactobacillus strains arrayed on MRS agar for antibacterial activity testing.
Photograph courtesy of Wilmar International Limited.
Are you lacking the power to get your
screening workflow over the line?
Our precision robots can help.