Tom Drake, Singer Instruments, Roadwater, UK.
Introduction
Colony picking is a vital part of many standard laboratory workflows. With higher throughputs, laboratories now consider automation to standardise this task. Colony pickers are required to consistently produce viable results, whilst tracking their actions for repeatability. This research investigates the efficiency of PIXL; a measure of the viable colonies that grow after picking.
Results
Utilising a standard laboratory setup, the efficiency of PIXL was investigated. Efficiency was defined as the average percentage of successful picks (hits), that then successfully formed colonies after an incubation period. The investigation used both Saccharomyces cerevisiae and Escherichia coli (strain information is detailed in Supplementary Figure 1), across three varieties of source plates that PIXL accepts; PlusPlates (SBS format agar plates), 90mm petri dishes and 150mm petri dishes.
The average percentage of colonies that grew after picking is shown, by plate type and organism, in Figure 1. Over a total of 9,690 picked colonies, 9,675 successfully grew after being picked with PIXL, giving a total average efficiency of 99.78%, with further data shown in Table 1.
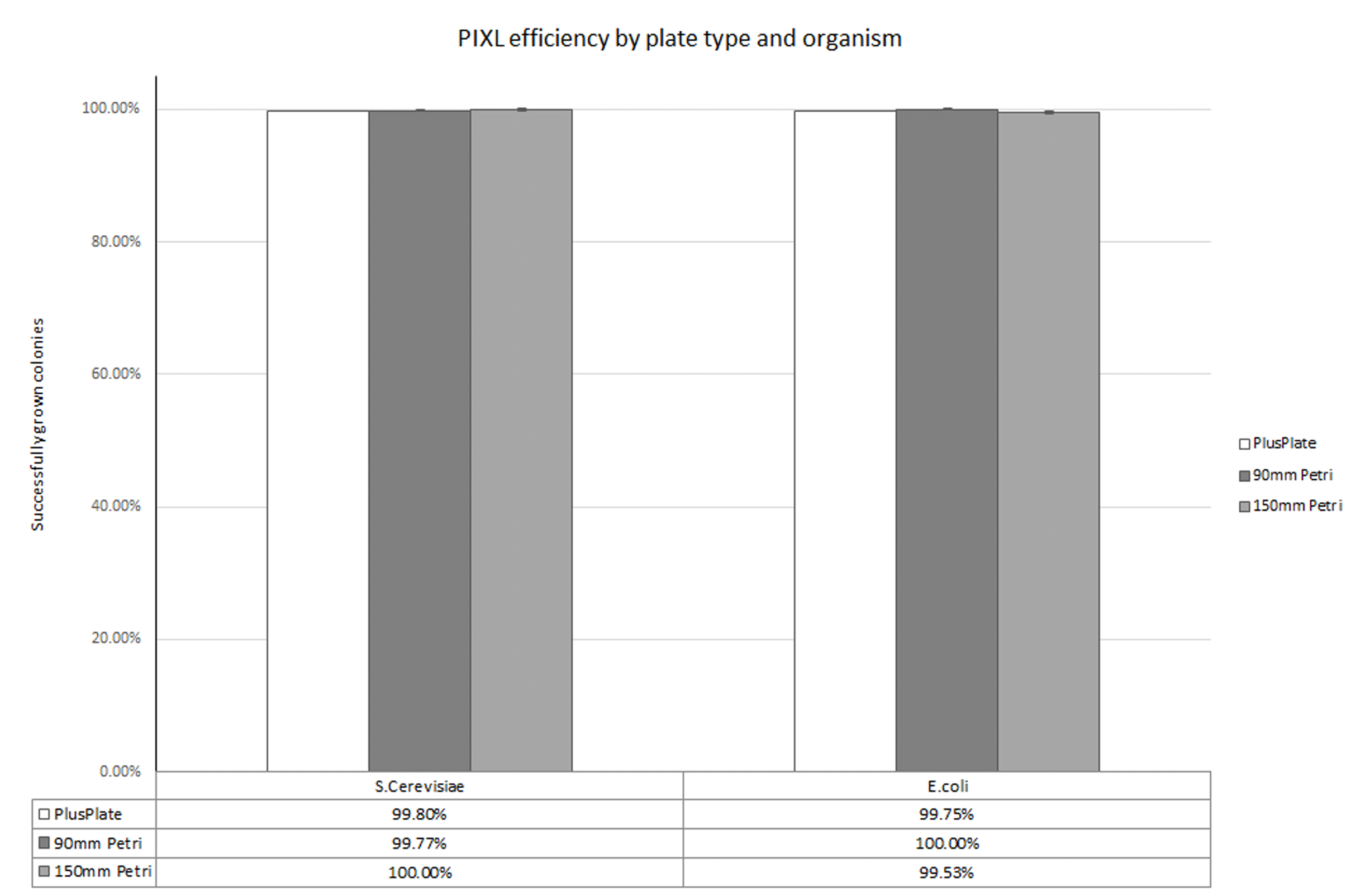
Fig 1. (Click to enlarge image)
Figure 1. PIXL efficiency by plate type and organism. The proportion of successfully gown colonies is shown for each strain and plate type, with final values highlighted under the figure. Error bars shown represent standard error and are not shown for values over 100%.
| Organism | Number Picked | Number Grown | Efficiency |
| S. cerevisiae | 7352 | 7345 | 99.90% |
| E. coli | 2338 | 2330 | 99.66% |
| Total | 9690 | 9675 | 99.78% |
Table 1. Raw efficiency data for all 9,690 colonies picked by PIXL for both tested strains, with final efficiency values. The total efficiency for PIXL is generated from the sum of grown/picked colonies, to account for the distribution of colonies by strain.
Discussion
The investigation measured efficiency; and successful growth after picking. This is a viable method to measure efficiency as this replicates the process within the laboratory, and also factors the repeatability, or precision, of PIXL. However, this method also integrates any error where colonies fail to grow as a result of other, external factors, into the measure of efficiency. Without taking the biological limitations into account, the total efficiency is 99.78%, as shown in Table 1. Hypothetically, this could be higher if the impact of any biological limitations were defined and accounted for.
A total of 9,690 colonies were picked across both yeast and bacterial samples, over three varieties of plate (including two different media). There were no marked differences between plate types, or strains, producing a very high overall efficiency, with only 15 colonies failing to grow out of 9,690. PIXL pins into arrays enabling easy manual identification of colonies which failed to grow.
Investigations into further aspects of PIXL are ongoing, and this data will be reported as it can be made publicly available.
Method
A serial dilution was produced for each strain from a 10 ml PBS suspension. Plated dilutions were used to calculate a CFU/ml to identify the volume required to isolate single colonies. At least 10 plates of random colonies were created for each strain and plate type. Plates were incubated at 30°C, for 48 hours for S. cerevisiae, and 24 hours for E. coli. Colonies were picked using default PIXL settings, onto PlusPlates containing YPD or LB (Formedium, UK) depending on organism. Plates were then incubated as before, and finally manually counted.
Final figures were calculated for each strain, and subset plate types as a percentage of viable colonies. Care was taken to not calculate an average from averaged results to minimise the error.
Discover how PIXL’s precision picking technology performs when picking non-model organisms in this article.
Explore the significance behind PIXL’s pinning pressure
Size Speed isn’t everything. PIXL’s Pinpoint picking technology eliminates contamination from cell splashing caused by high-speed pinning approaches.