- Products
- Resources
Singer Instruments at the 8th International Conference on Microbial Diversity!
When in Rome… come and see our instruments in action!
September 23rd-26th 2025
Singer Instruments presenting at the 8th International Conference on Microbial Diversity
We’re proud and excited to announce that this year, Singer Instruments are making their way to Rome for the 8th International Conference on Microbial Diversity, taking place 23-26th September 2025.
The conference is bringing together researchers from diverse backgrounds to share findings, resources and methods to propel and scale microbial technologies for a broader impact.
Aside from advances in the functional studies of soil microbiome, the conference will also cover the prospects and challenges of scaling research into microbes and microbe-derived natural products across a range of globally relevant themes such as food systems security, solutions for sustainable agriculture and waste management.
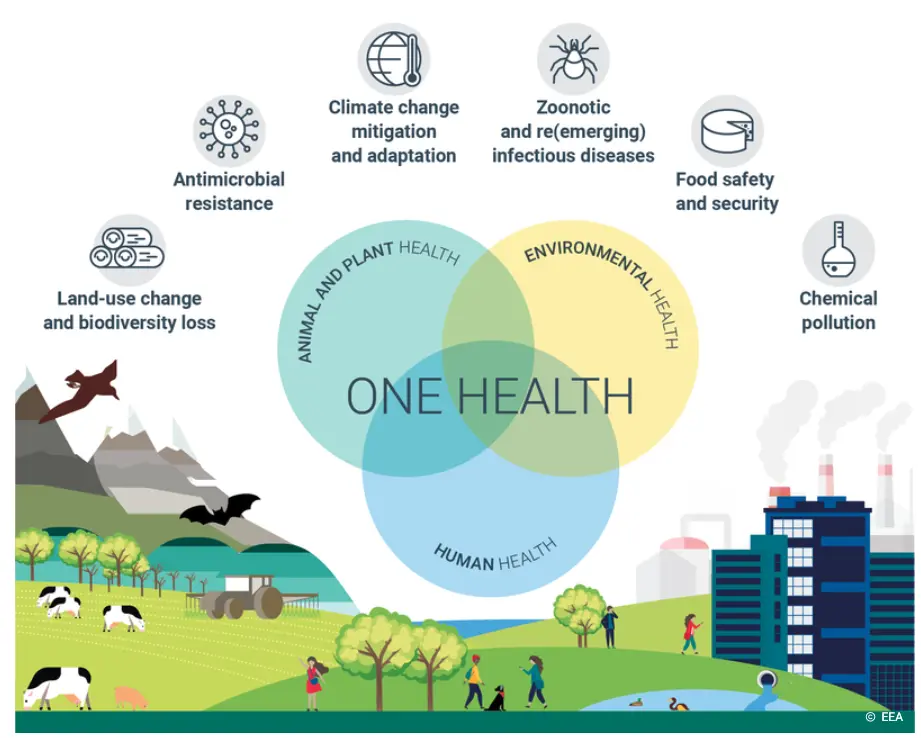
With our longstanding track record in manipulating, replicating and picking diverse microorganisms, we wanted to share some of the ways researchers are leveraging our screening and picking technologies to tackle complex challenges of the ‘One Health’ approach (Figure 1).
Sourcing soil derived Corynebacterium sp. isolate that outperforms established strains using ROTOR+
Lignocellulosic biomass (LCB) is a catchy name for the ubiquitous plant dry matter waste, rich in biodegradable polymers cellulose, hemi-cellulose and lignin (Figure 2). It’s a byproduct of several industrial processes in agriculture and forestry, such as saw and paper mill waste, sugarcane bagasse, and other plant residue (e.g. straw). Bioprocessing LCB yields various value-added products like biofuels, textile and adhesives, notably contributing to the circular economy (Haq et al., 2020).
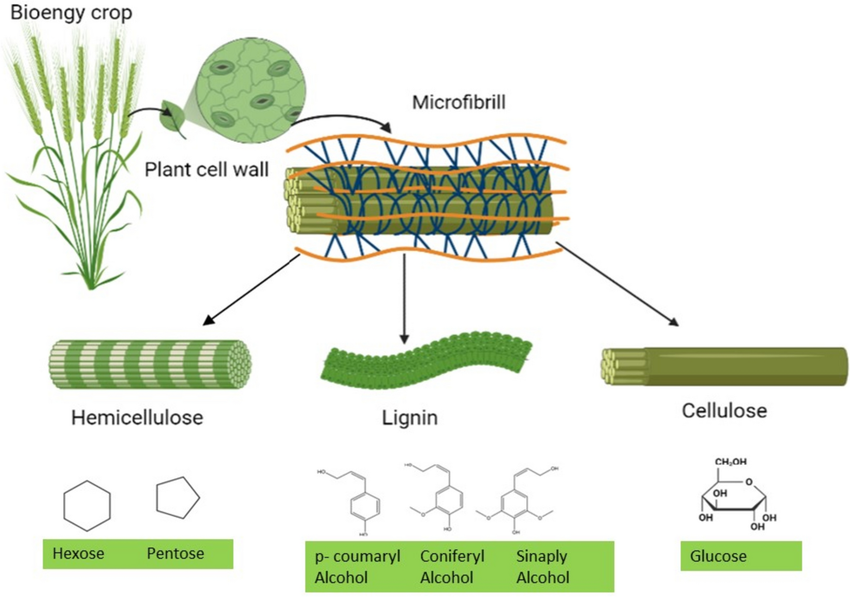
LCB must undergo pre-treatments like hydrolysis to release fermentable carbon sources. However, this process also releases known inhibitors to microbial growth.
Researchers from the Technical University of Denmark (Weiẞ et al., 2024) have developed a fast solid-media screening method using Singer Instruments’ ROTOR+ to measure colony fitness as a proxy for liquid-based screens.
The team followed a multi-step screening process:
- They screened and scored 32 strains to find the most suitable ones for LCB bioprocessing. This initial screening took place on agar plates containing various carbon sources.
- Next, they tested the strains’ tolerance to common inhibitors, including acetic acid, formic acid, furfural, 4-HBA, and vanillin.
- Finally, the top-performing strains were chosen for micro-fermentations. Their growth was directly scored in a medium of softwood spent sulfite liquor (SSL), a pulp and paper industry-derived LCB.
The study yielded Corynebacterium sp. ATCC 14747 as the top performer, surpassing Corynebacterium glutamicum, already characterised, engineered and utilised by the industry (Jin et al., 2020).
Rather than relying solely on synthetic biology to improve known strains, this approach leverages innate microbial phenome diversity to provide solutions for environmental sustainability.
Screening and phenotyping Australia’s S. cerevisiae genetic landscape with PIXL
The brewing of wine has been intertwined with humanity for thousands of years. Previously, spontaneous fermentation occurred with native species, resulting in distinct wine taste profiles confined to a specific geographical region. In modern practices, it is commonplace to use starter well-characterised monocultures which provide predictable and repeatable flavours and aromas.
The Australian Wine Research Institute (AWRI) has set out to describe the genetic diversity of Australian yeast species. They hypothesised feralisation of industrial strains and their admixture with other domesticated and non-domesticated lineages present in Australia, itself presumed scarce in endemic strains (Ward et al., 2024).
The researchers isolated 411 S. cerevisiae strains from spontaneous ferments in a high-throughput manner, utilising Singer’s PIXL for single colony picking. This was followed by genome mapping, as well as a detailed phylogenetic analysis.
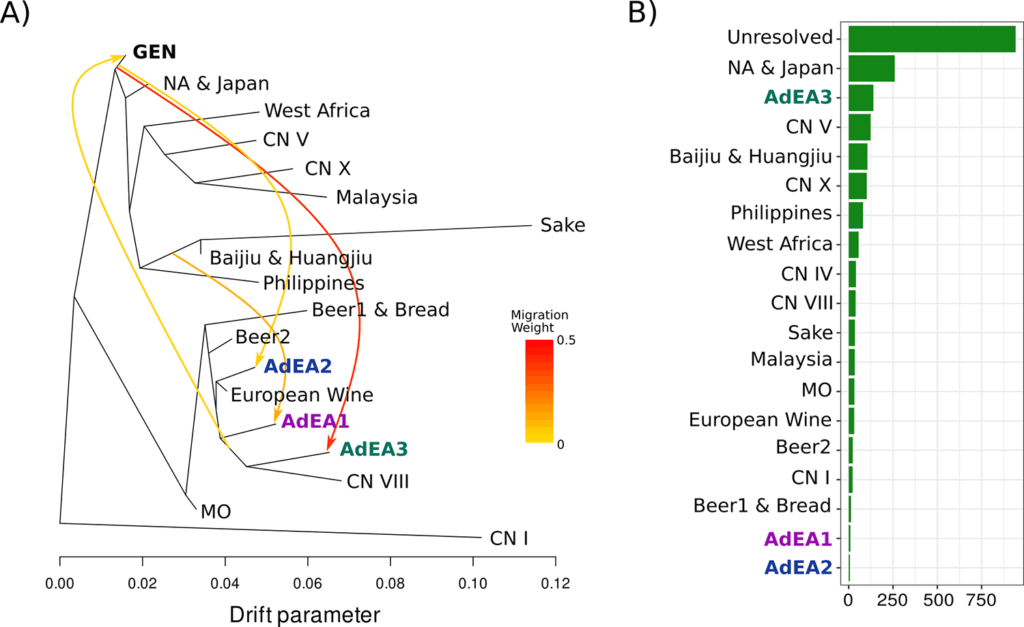
The results show that Australia’s S. cerevisiae population landscape is the product of a highly recombined mix of multiple lineages related to the curated strains of the Wine Reference Panel (WRP).
They also found evidence of a widely spread, but non-domesticated, non-European strain that may predate European colonisation. These findings open up questions about how commercial strains shape the genetics of native yeast biodiversity and raise questions about the impact of loss of microbial terroir (Figure 3).
Screening S. cerevisiae to uncover sodium bicarbonate stress responses using ROTOR+ and Phenobooth+
It’s estimated that over a billion hectares of land are affected by excess soil salination and are therefore unsuitable for crop cultivation (Cao et al., 2022). Whether occurring naturally from weathering or induced by irrigation (with excess salts like sodium bicarbonate and sodium carbonate), saline-alkaline soil negatively impacts both plant and microbial growth.
The cellular pathways conferring resistance to sodium bicarbonate (NaHCO3) stress, as well as the mode of its antimicrobial activity remain unclear. Researchers from Zhejiang A&F University in Hangzhou, China set out to elucidate these genetic factors using synthetic genetic arrays (SGA) in S. cerevisiae BY4741 (Cao et al., 2022).
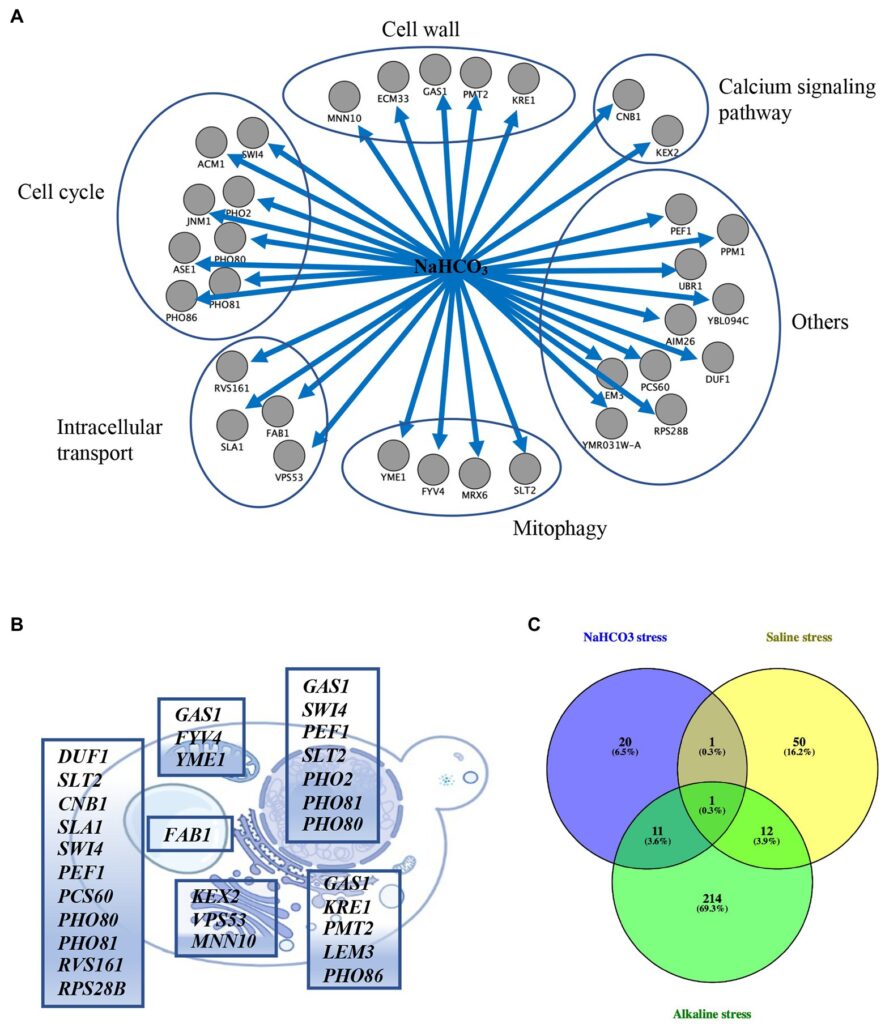
They screened the collection of 4200 single-gene deletion mutants in presence and absence of NaHCO3 using ROTOR+ to facilitate the screens and PhenoBooth+ to quantify the responses. Using our instruments they identified:
- 33 genes directly responding to NaHCO3 stress.
- 309 upregulated and 233 downregulated genes (Figure 4).
- NaHCO3 stress response that is distinct from other saline and alkaline conditions.
The findings from this study has far reaching applications; helping us to understand:
- Stress responses to salinated soil.
- How oral microbes respond to NaHCO3– a common additive in toothpaste.
- NaHCO3‘s applications in argiculture where it is a common additive in animal feed and post-harvest fungicides.
This research, therefore, sets out a clear path to study NaHCO3 stress and how microorganisms adapt to cope with its toxicity. More broadly, genome-wide screening is an effective methodology to understanding response mechanisms in yeasts and beyond.