- Products
- Resources
Passion brought us here. And yes yeast. Lots of yeast!
As we gear up for ICYGMB32 (the 32nd International Conference on Yeast Genetics and Molecular Biology, July 21-24 2025) later this month, we’ve been thinking a lot about how yeast has shaped the history of Singer Instruments.
Our journey with yeast stems all the way back to the 1950’s and early experiments to separate the spores of a tetrad and analyse their progeny. This is really what led to Singer Instruments becoming somewhat of a household name within the yeast community (it turns out more than 60% of invited speakers are already our customers)!
The truth is that over the years, your methodological challenges and hiccups have become the near obsession of our engineers and scientists. And it’s largely through the generous insights (and particularly good humour) of the yeast community that Singer Instruments’ wide range of microbe-inspired products have been entwined with yeast biology for so long.

Today our industry-leading microbial workstations have more than 10,000 citations and counting! As we raise a glass to Singer Instruments’ 90+ years of service to science (the company was founded in 1934), we wanted to shine a light on just some of those outstanding contributions from recent years.
So sit tight as we take you through our “Singer Stars of the Yeast Conference.”✨
1) Paul Nurse
Nobel Laureate and MSM 400 tetrad dissection pioneer

Paul was a good friend of the late Carl Singer. In 1989 he became one of the first ever customers for our infamous MSM tetrad dissection microscope. Paul says he first learned tetrad dissection in the lab of Urs Leupold at the University of Bern in Switzerland, back in the late 70’s when it was traditional to dissect by hand, without a micromanipulator!
But since discovering the MSM 400, he’s been confident to hand over the task to just about anyone in the lab:
“Basically I can just say ‘Look, here is the machine. There is the manual, get on with it, play with it and work.’ The MSM 400 really has been quite an important and useful addition to our lab,” he told Singers during a visit to The Crick in 2017.
Much of Paul’s Nobel Prize winning work is based on the model organism Schizosaccharomyces pombe, “pombe” being the Swahili for beer. “The taste was sweet and very alcoholic but definitely drinkable” Paul wrote upon returning from a visit to a Ugandan brewery in 2016 to trace its origins (Vyas & Freitas, 2022).
If you can’t dissect it, drink it. Or so goes the old saying.
2) Michael Chang
Group Leader at the European Research Institute for the Biology of Ageing (ERIBA) and ROTOR+ SGA black belt

Having undertaken his PhD in Grant Brown’s lab at the University of Toronto and later spent time in Rodney Rothstein’s lab at Columbia University Medical Center as a post doc, Michael is well-versed in the art of Synthetic Genetic Array.
SGA is a technique that was rapidly gaining in traction around the time our ROTOR+ colony pinning robot launched twenty years ago.
ROTOR+ subsequently largely replaced liquid-based approaches to the screening of large microbial libraries, due to its ability to accurately pin colonies on agar at densities of up to 6144 (Bean et al, 2014). That’s the entire S. cerevisiae deletion library onto a single SBS plate!
At the helm of his own lab based at ERIBA , his team are now applying the technique to dissect the cellular mechanisms that maintain genome integrity and telomere function (Rosas Bringas et al, 2024).
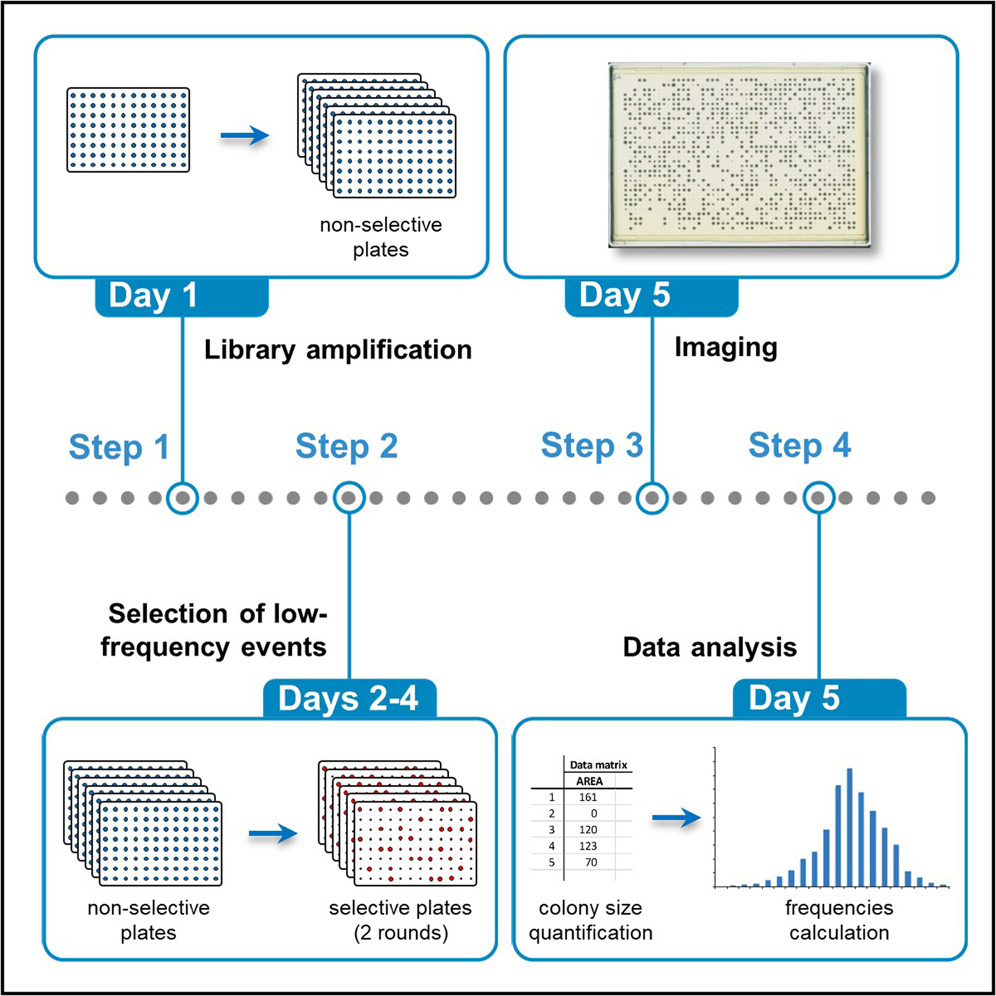
Michael’s experience with SGA and ROTOR+ has notably contributed to establishment of a 5-day high-throughput approach to screening for yeast genes controlling low-frequency events (Novarina et al, 2022).
Michael and colleagues have successfully employed the protocol in genome-wide screens to identify genes that suppress the accumulation of spontaneous mutations in young and aged yeast cells (Novarina et al, 2019) and also that affect spontaneous direct-repeat recombination, a key driver of cancer (Novarina et al, 2020).
3) Elena Kuzmin
Assistant Professor in Synthetic and Functional Genomics and winner of the 2012 Singer Tetrad Dissection Olympics
Elena was once described as “a role model for young women in science” after being awarded a L’Oréal Canada Women in Science Research Excellence Fellowship in 2019. We enjoyed witnessing Elena’s talents firsthand, crowning her our gold medalist at Singer Instruments’ inaugural Yeast Tetrad Dissection Olympiad in 2012! Even by today’s standards, dissecting five tetrads in 5 minutes and 22 seconds takes some serious beating.
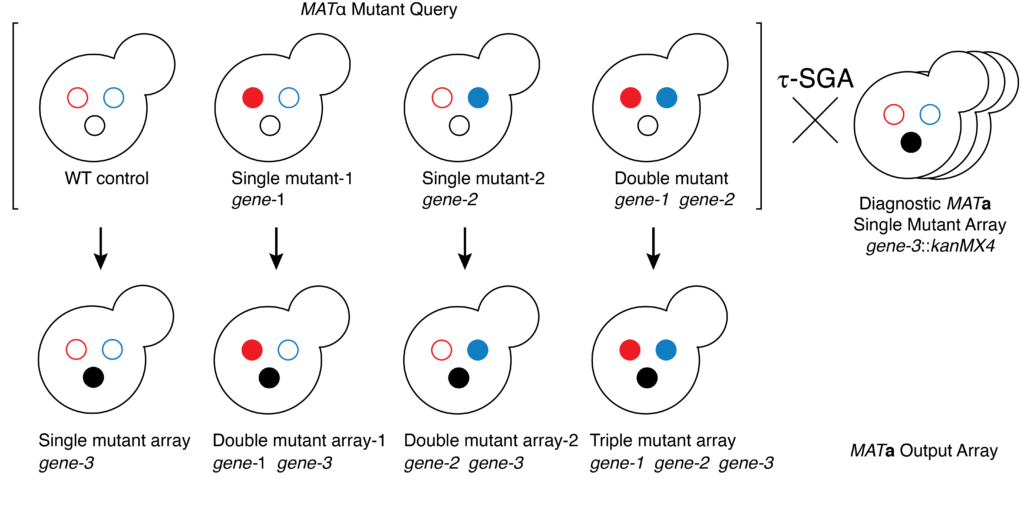
Following her Fellowship, Elena wasted no time in establishing her own research program, building on pivotal work she led while at the University of Toronto (under the supervision of Professors Charlie Boone and Brenda Andrews) to create a process for mapping trigenic genetic interactions in yeast using synthetic genetic array, (Kuzmin et al, 2021).

Channeling her experience with ROTOR+ and also SporePlay+, Elena went on in 2020 to front a systematic analysis of the evolutionary trajectories of duplicated genes, helping to shed light on the mysterious persistence of functional redundancy within genomes (Kuzmin et al, 2020).
Elena now divides her time between Concordia University’s Applied Synthetic Biology and Structural and Functional Genomics Centres, and McGill University’s Rosalind & Morris Goodman Cancer Institute, where her team is focused on applying her high-throughput approaches to progress understanding of human health and disease.
4) Joseph Schacherer
Founder of the 1002 genomes project, with a little help from the ROTOR+ PIXL high throughput screening combo

We’re grateful to Joseph for the kind invitation to exhibit at ICYGMB32. Our last planned rendezvous with him was on a visit to our premises, hot off the heels of the Yeast 1002 Genome project’s 2018 Nature paper, marking the culmination of an unprecedented project to sequence and phenotype the genomes of 1,011 Saccharomyces cerevisiae isolates from around the world.
This monumental task provided the most comprehensive genomic and phenotypic dataset of the species to date. Facilitated by our PIXL and ROTOR+ robots, the project has profoundly impacted the field of yeast genetics and beyond, providing the foundational dataset for countless subsequent studies.
Among some of the latest publications from Joseph’s acclaimed lab, we spotted a compelling series of studies tackling the fundamental question of how an organism’s genes create its traits by mapping different layers of gene regulation. Their work on the first layer was detailed in a major Nature Genetics study, where the team charted the genetic basis of RNA expression across more than a thousand yeast isolates (Schacherer et al., 2024). This created a foundational map of the yeast transcriptome, setting the stage for the next critical question: does the same genetic logic apply to proteins?
To answer this, a subsequent PNAS study, led by the Schacherer lab and in collaboration with Markus Ralser’s team, tackled the protein landscape (Schacherer et al., 2024). We’re again proud of the small but vital part of ROTOR+, providing the necessary grunt to generate precise proteomic strain libraries for 942 diverse natural isolates of S. cerevisiae. This work led to a crucial discovery: the genetic rules that control protein abundance are almost entirely separate from those for RNA. But perhaps even more importantly, this study serves as a potent reminder that what is seen in one genetic background, no matter how meticulously analysed, may not be universally representative.
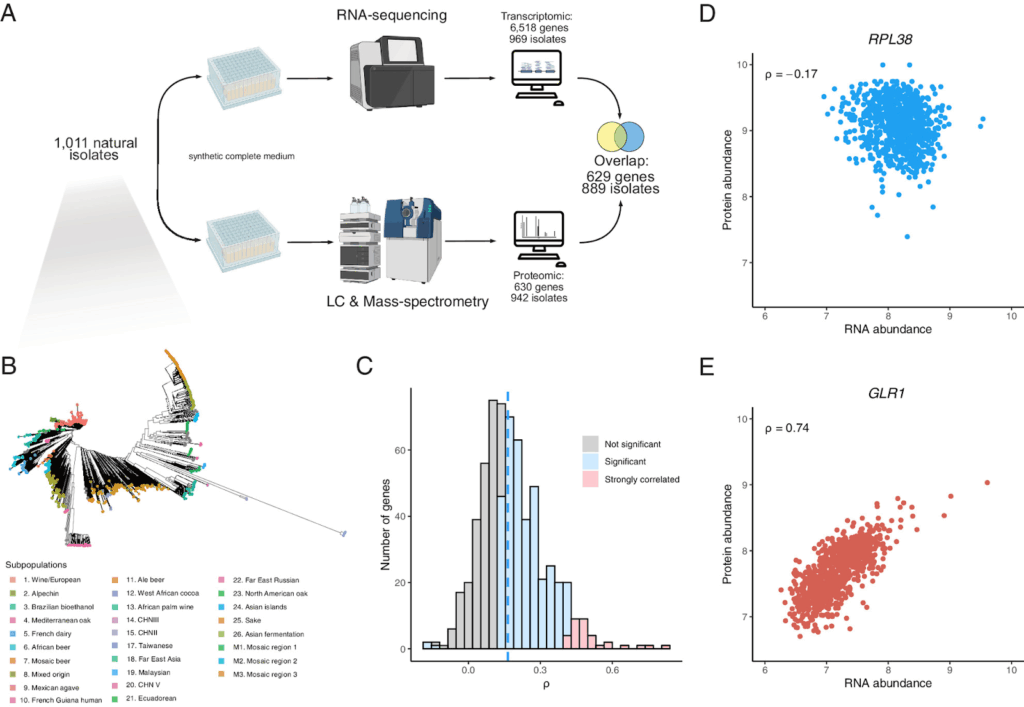
Quantitative proteomes and transcriptomes of a large S. cerevisiae population. (Schacherer et al., 2024).
See you at the conference!
It’s true, our customers are publishing far too much exciting research to cover in a single blog post. But we’ll have many more interesting studies you can browse on our stand at ICYGMB32. Do come and say ‘hi’. We’d love to geek out with you about yeast and perhaps reacquaint you with our robot overlords.
