Aim
To determine whether PIXL can pick and plate a range of non-model organisms including a filamentous fungus and a range of actinobacteria.
Introduction
Microorganisms represent a major source of chemical diversity and the rate at which this huge resource can be screened is a limiting factor to the discovery of new compounds including chemical feedstocks, antibiotics, and other pharmaceuticals. New high-throughput screening processes must be developed in order to be able to efficiently exploit the vast microbial resources that are available.
PIXL, a new precision colony-picking robot, addresses this bioprospecting challenge, by automating some of the culturing and downstream processing necessary for high-throughput experimentation with these organisms. PIXL has a proven transfer efficiency of >99% using common model organisms including E. coli and S. cerevisiae, but to understand PIXL’s full capability the team at Isomerase, a Cambridge UK-based biological engineering company, tried to ‘beat’ PIXL with a range of organisms that they work with, some of which they were confident would trip PIXL up. Here is the background to the challenge.
One of PIXL’s core functions is to be able to automatically detect colonies and then pick these to new plates for culturing or downstream analysis. One challenge posed by the diversity of microorganisms is that they look and behave differently. For a colony-picking robot, detection algorithms are challenged by colonies that have unusual colours or morphologies. Furthermore, if the physical properties of the cells prohibit adherence to the picking device, or the cells do not survive being picked, then it will prove difficult to effectively automate cell transfer.
Isomerase were interested to understand whether PIXL could pick colonies of species that exhibited atypical phenotypes. This included a dark brown filamentous fungus and a wide range of actinomycetes from around the world. The organisms chosen were not only different colours but also exhibited different colony morphology, from dry and powdery, to extremely tenacious. How did PIXL perform against these challenges?

Fig 1. Collection of Isomerase source plates picked using PIXL.
Results
This is what Martin Sim from Isomerase had to say about how PIXL performed:
“I’m happy to say that every strain we picked and replica plated onto the solid agar in the 96 well plates grew, and after three weeks in the incubator no plate has shown evidence of contamination”
The advantage of being able to automatically pick and plate colonies is clear and PIXL’s success demonstrates how sophisticated this colony-picking robot is. But what is it like ‘in the field’? Can it handle uneven agar surfaces? Sophisticated can sometimes mean complicated, so is PIXL difficult to work with? Here is Martin again discussing how he and his team found working with PIXL:
“Everyone was very impressed with the machine, especially its ease of use and the friendly software user interface, the polymer picking system, and of course how it was accurately handling uneven agar surfaces, and working with our unusual colony morphologies.”
Conclusion and Benefits
PIXL successfully detected and picked a variety of fascinating organisms chosen by Isomerase. PIXL has already been shown to detect and pick model organisms such as S. cerevisiae (yeast), E. coli (gram-negative bacteria) and C. reinhardtii (algae). The data reported here demonstrate that PIXL is also capable of working with non-model organisms that exhibit atypical and challenging phenotypes. In light of this, PIXL could be readily utilised in bioprospecting applications as well as in other fields that use non-model organisms.
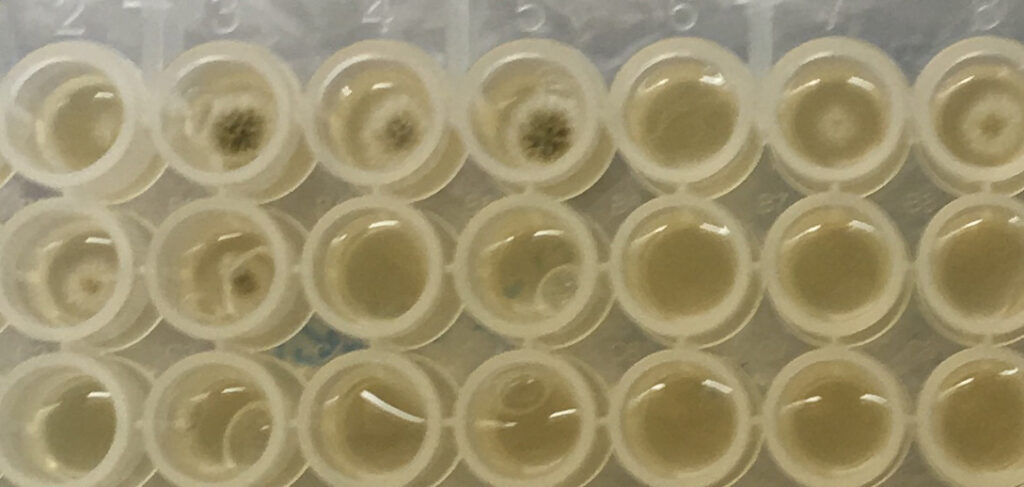
Glarea lozoyensis
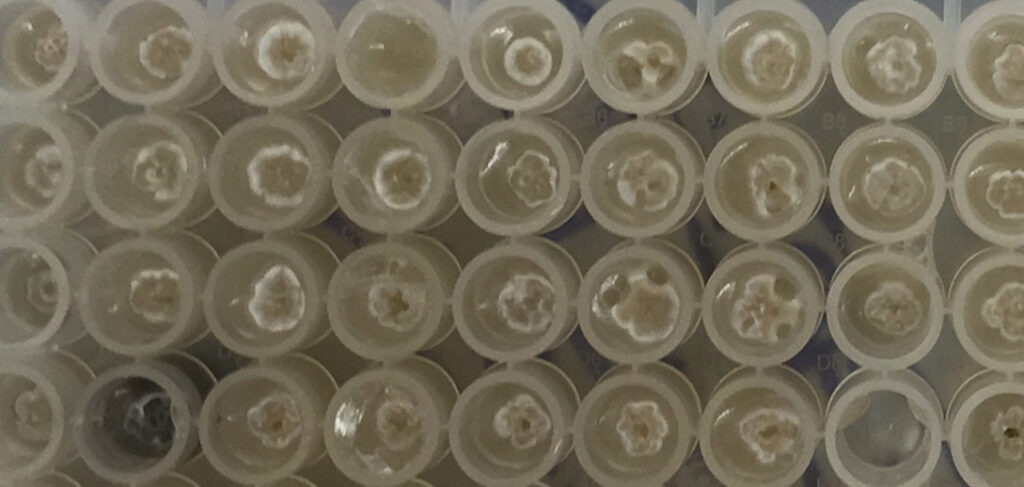
Kitasatospora putterlickiae
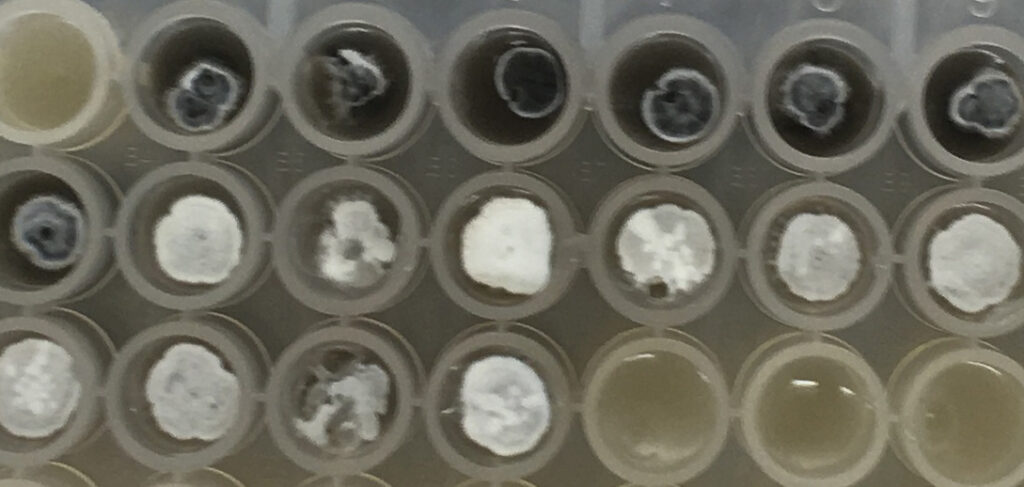
Streptomyces avermitilis & streptomyces venezuelae
Figure 2. PIXL picked colonies – PIXL successfully picked colonies of different species chosen by Isomerase. The species chosen exhibited a range of morphologies that PIXL was able to detect, pick and re-plate.
Do you want to reliably pick non-model organisms?
Get in touch with our Product Experts to learn how PIXL can automate your workflow.