What microorganisms can PIXL pick?
A roundup of the latest evidence
Can you pick it? Yes you can
Microorganisms come in many funky shapes, sizes, and textures, from the colourful and gloopy to the funky and filamentous. Since PIXL’s launch in 2017, our users have tried and tested the instrument with a wide variety of different model and non-model organisms.
PIXL achieves consistently high transfer rates for E. coli and S. cerevisiae, which is perhaps unsurprising since these are the two main microorganisms it was first engineered to pick. But many of our customers are now increasingly diversifying into a wide variety of other organisms.
Here’s a roundup of some of the latest studies demonstrating PIXL’s prowess across a range of different morphologies.
Escherichia coli
It’s a cornerstone of biomedical research: Escherichia coli has been somewhat the bacterial model organism of choice since the 1940’s and remains popular among PIXL users today.
Scientists based at the Stanford University Department of Bioengineering, for example, have been using PIXL to delve into the world of mutant E. coli. Using expansive 15 cm source petri dishes teeming with up to 500 colonies per plate, they have made use of PIXL’s high throughput screening abilities to help filter and transfer colonies of interest into 384 multi-well plates. This has contributed to the development of an efficient method for preparing protein mutant libraries and understanding their biochemistry (Appel et. al., 2021).
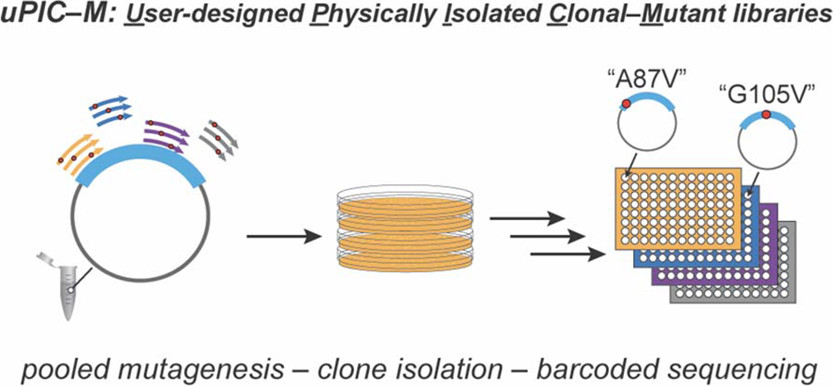
Figure 1: Stanford University scientists have used PIXL to support development of a scalable, rapid, and inexpensive approach for creating clonal-mutant libraries. Image source: ACS Omega 2021, 6, 45, 30542-30554. doi.org/10.1021/acsomega.1c04180
Saccharomyces cerevisiae
Together with the MSM 400, PIXL has helped yeast researchers working with S. cerevisiae pioneer the high-throughput investigation of colonies formed by dissected tetrads.
This follows many decades of Singer Instruments collaborating with the yeast genetics community to develop precision technologies for the micromanipulation of individual yeast cells (Singer et al, 2021).
For example, PIXL was used in research led by Professor Joseph Schacherer at the University of Strasbourg, helping to evaluate variation in mutant fitness across different environments among 106 natural isolates selected from the S. cerevisiae 1,011 genomes collection (Caudal et. al., 2022).
Such research is helping scientists to uncover how identical mutations can produce different phenotypes depending on the genetic background, a phenomenon also observed in many human diseases.
Wild yeasts
While lab grown strains of baker’s yeast remain a mainstay of microbiology, there is also growing interest in non‐Saccharomyces yeast strains, not least within the fermentation industry.
For example, researchers at the Manchester Institute of Biotechnology in collaboration with Singer Instruments successfully used our high throughput screening suite to analyse some of the best yeasts for cider fermentation.
Crucially, they were able to use PIXL to successfully explore a wide range of morphological phenotypes including strains isolated from worldwide breweries, Hawaiian trees, German Cockroaches and Illinois’ Cream (Aguiar-Cervera et. al., 2021).
The team used PIXL to re-array a library of six conventional and 17 non‐conventional yeast strains into 96, 384 and 1536 arrays onto solid agar. They then employed ROTOR+ to replicate the library in various industrially-relevant conditions, followed by Phenobooth+ to obtain colony radius values measured in pixels for all strains and timepoints.
It’s hoped the method could be used to dramatically increase throughput for a range of industrial applications involving the screening vast yeast libraries formed by unknown isolates (see figure 1).
For more tips on automating high throughput screening, library construction and strain optimisation, read Taking the strain out of working with strains.
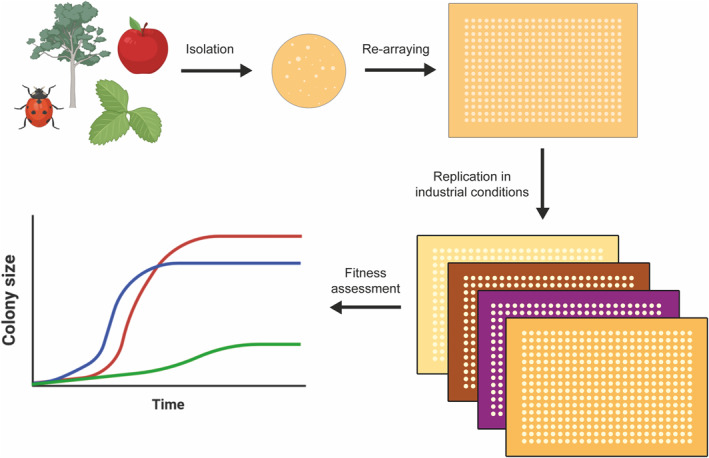
Figure 1: Using ROTOR+ PIXL, researchers at the Manchester Institute of Biotechnology collaborated with Singer Instruments to re-array and replicate a vast number of Saccharomyces and non‐Saccharomyces isolates in solidified media representing industrial conditions. Image source: Eng Biol. 2021 Aug 30;5(3):72–80. doi: 10.1049/enb2.12013.
Other prokaryotes
Work in collaboration with Shimadzu to develop PIXL for automated MALDI-TOF sample preparation has also provided evidence for PIXL’s ability to pick non-Escherichia prokaryotic organisms, including Bacillus subtilis, Cellulomonas uda, and Pantoea agglomerans.
This has resulted in a new protocol that supports more accurate bacterial identification through improving the consistency and throughput of transferring unknown bacterial colonies onto MALDI-TOF plates.

Chlamydomonas reinhardtii
The eukaryotic alga Chlamydomonas reinhardtii has become firmly established in recent years as one of the best microbial genetic models for the plant superkingdom. However, the gloopy green morphology of Chlamydomonas differs markedly from budding yeast, not just in morphology but also in cell size, growth rate, colony adhesiveness and light responsiveness. How might PIXL perform?
Professor Kyle Lauersen’s lab at the King Abdullah University of Science and Technology (KAUST) should know. They are successfully using PIXL in combination with ROTOR+ to develop a variety of resource-efficient bio-processes involving genetically engineered algae.
In fact, PIXL’s ability to screen colonies for red or yellow fluorescent protein expression has been a component in several studies by the team aimed at strain optimisation for isoprene production (Abdallah et. al., 2022; Yahya et. al., 2023).
Through the genetic modification of green algae, this research, and by implication PIXL, is thereby supporting efforts to meet the market demand for rubber in an environmentally friendly manner.

Filamentous fungi
At Singer Instruments, we are always keen to test our products with new organisms. So we couldn’t resist accepting the challenge when two different bioprospecting companies offered to test how PIXL would perform when picking various filamentous fungi.
Among the test organisms chosen by Isomerase was the dark brown filamentous fungus Glarea Lozoyensis, which is of medical interest for its antifungal properties. PIXL proved it could pick this and more with no evidence of contamination.
PIXL has further undergone preliminary user testing with Aspergillus Niger and Aspergillus Nidulans. Using its unique ability to ‘dry mix’ with zero vertical travel (essentially dragging the picking head gently through the fungal colony), PIXL was successfully demonstrated to pick all but two of the 56 test colonies (see figure 2). Further controlled tests are required to generate more data but these preliminary results are very encouraging.
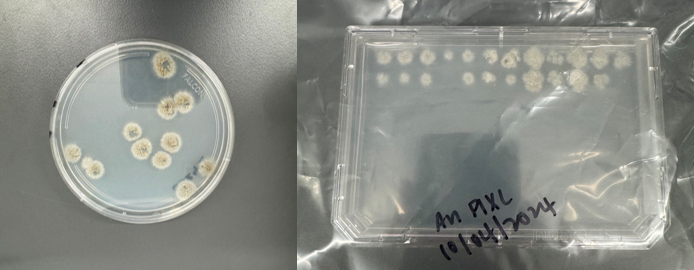
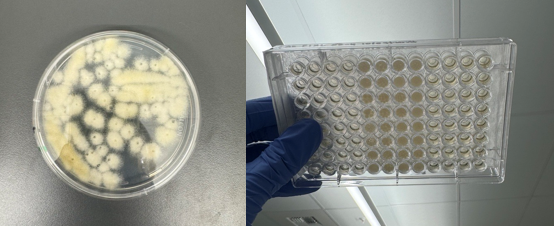
Figure 2: PIXL’s unique ability to ‘dry mix’ facilitates the picking of Aspergillus and other filamentous species.
Summary
This is by no means an exhaustive list of PIXL success stories, but we hope it gives you a flavour of the versatility of this instrument. In fact, our experience shows that if you can pick it by hand (without too much fuss or digging), you can be fairly confident that PIXL can pick your colonies too. Why not challenge us to put it to the test?
Looking for lab automation with flexibility that scales with you?
PIXL: the only colony picker capable of seamlessly handling
multiple organisms without missing a beat.

By Fiona Kemm | Research Scientist
Fiona plays a key role in ensuring our innovations perform as intended, while providing exceptional support to users. Her efforts in the lab contribute to the reliability of our robots, developing protocols and assays that demonstrate their unique capabilities. She has a BSc in Biochemistry, an MRes in Molecular Microbiology and a talent for BioArt.